Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces
- PMID: 10773076
- PMCID: PMC105376
- DOI: 10.1093/nar/28.10.2084
Codon usage in Chlamydia trachomatis is the result of strand-specific mutational biases and a complex pattern of selective forces
Abstract
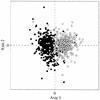
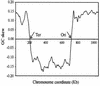
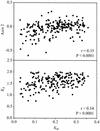
The patterns of synonymous codon choices of the completely sequenced genome of the bacterium Chlamydia trachomatis were analysed. We found that the most important source of variation among the genes results from whether the sequence is located on the leading or lagging strand of replication, resulting in an over representation of G or C, respectively. This can be explained by different mutational biases associated to the different enzymes that replicate each strand. Next we found that most highly expressed sequences are located on the leading strand of replication. From this result, replicational-transcriptional selection can be invoked. Then, when the genes located on the leading strand are studied separately, the correspondence analysis detects a principal trend which discriminates between lowly and highly expressed sequences, the latter displaying a different codon usage pattern than the former, suggesting selection for translation, which is reinforced by the fact that Ks values between orthologous sequences from C. trachomatis and Chlamydia pneumoniae are much smaller in highly expressed genes. Finally, synonymous codon choices appear to be influenced by the hydropathy of each encoded protein and by the degree of amino acid conservation. Therefore, synonymous codon usage in C.trachomatis seems to be the result of a very complex balance among different factors, which rises the problem of whether the forces driving codon usage patterns among microorganisms are rather more complex than generally accepted.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

