Supercomplexes in the respiratory chains of yeast and mammalian mitochondria
- PMID: 10775262
- PMCID: PMC302020
- DOI: 10.1093/emboj/19.8.1777
Supercomplexes in the respiratory chains of yeast and mammalian mitochondria
Abstract
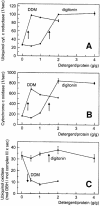
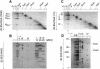
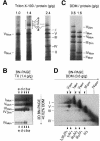
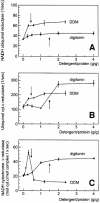
Around 30-40 years after the first isolation of the five complexes of oxidative phosphorylation from mammalian mitochondria, we present data that fundamentally change the paradigm of how the yeast and mammalian system of oxidative phosphorylation is organized. The complexes are not randomly distributed within the inner mitochondrial membrane, but assemble into supramolecular structures. We show that all cytochrome c oxidase (complex IV) of Saccharomyces cerevisiae is bound to cytochrome c reductase (complex III), which exists in three forms: the free dimer, and two supercomplexes comprising an additional one or two complex IV monomers. The distribution between these forms varies with growth conditions. In mammalian mitochondria, almost all complex I is assembled into supercomplexes comprising complexes I and III and up to four copies of complex IV, which guided us to present a model for a network of respiratory chain complexes: a 'respirasome'. A fraction of total bovine ATP synthase (complex V) was isolated in dimeric form, suggesting that a dimeric state is not limited to S.cerevisiae, but also exists in mammalian mitochondria.
Figures



References
-
- Anemüller S., Lübben,M. and Schäfer,G. (1985) The respiratory system of Sulfolobus acidocaldarius, a thermoacidophilic archaebacterium. FEBS Lett., 193, 83–87. - PubMed
-
- Berry E.A. and Trumpower,B.L. (1985) Isolation of ubiquinol oxidase from Paracoccus denitrificans and resolution into cytochrome bc1 and cytochrome c–aa3 complexes. J. Biol. Chem., 260, 2458–2467. - PubMed
-
- Boumans H., Grivell,L.A. and Berden,J.A. (1998) The respiratory chain in yeast behaves as a single functional unit. J. Biol. Chem., 273, 4872–4877. - PubMed
-
- Bruel C., Brasseur,R. and Trumpower,B.L. (1996) Subunit 8 of the Saccharomyces cerevisiae cytochrome bc1 complex interacts with succinate–ubiquinone reductase complex. J. Bioenerg. Biomembr., 28, 59–68. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

