The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast
- PMID: 10775265
- PMCID: PMC302011
- DOI: 10.1093/emboj/19.8.1803
The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast
Abstract
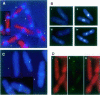
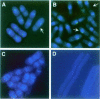
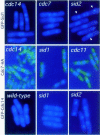
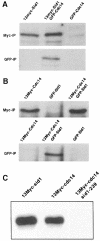
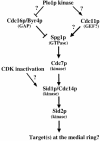
Coordination of mitosis and cytokinesis is crucial for ensuring proper chromosome segregation and genomic stability. In Schizosaccharomyces pombe, the sid genes (cdc7, cdc11, cdc14, spg1, sid1, sid2 and sid4) define a signaling pathway that regulates septation and cytokinesis. Here we describe the characterization of a novel protein kinase, Sid1p. Sid1p localizes asymmetrically to one spindle pole body (SPB) in anaphase. Sid1p localization is maintained during medial ring constriction and septum synthesis and disappears prior to cell separation. Additionally, we found that Cdc14p is in a complex with Sid1p. Epistasis analysis places Sid1p-Cdc14p downstream of Spg1p-Cdc7p but upstream of Sid2p. Finally, we show that cyclin proteolysis during mitosis is unaffected by inactivating the sid pathway; in fact, loss of Cdc2-cyclin activity promotes Sid1p-Cdc14p association with the SPB, possibly providing a mechanism that couples cytokinesis with mitotic exit.
Figures

Similar articles
-
Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis.J Cell Biol. 1999 Aug 23;146(4):777-90. doi: 10.1083/jcb.146.4.777. J Cell Biol. 1999. PMID: 10459013 Free PMC article.
-
Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast.Curr Biol. 2000 May 18;10(10):619-22. doi: 10.1016/s0960-9822(00)00492-9. Curr Biol. 2000. PMID: 10837231
-
Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression.Curr Biol. 2001 Jun 26;11(12):931-40. doi: 10.1016/s0960-9822(01)00268-8. Curr Biol. 2001. PMID: 11448769
-
Closing mitosis: the functions of the Cdc14 phosphatase and its regulation.Annu Rev Genet. 2004;38:203-32. doi: 10.1146/annurev.genet.38.072902.093051. Annu Rev Genet. 2004. PMID: 15568976 Review.
-
Cell division: SIN, cytokinesis and ethanol dependency.Curr Biol. 2005 Aug 9;15(15):R605-7. doi: 10.1016/j.cub.2005.07.045. Curr Biol. 2005. PMID: 16085485 Review.
Cited by
-
Etd1p is a novel protein that links the SIN cascade with cytokinesis.EMBO J. 2005 Jul 6;24(13):2436-46. doi: 10.1038/sj.emboj.7600705. Epub 2005 Jun 2. EMBO J. 2005. PMID: 15933715 Free PMC article.
-
The nuclear kinase Lsk1p positively regulates the septation initiation network and promotes the successful completion of cytokinesis in response to perturbation of the actomyosin ring in Schizosaccharomyces pombe.Mol Biol Cell. 2005 Jan;16(1):358-71. doi: 10.1091/mbc.e04-06-0502. Epub 2004 Nov 10. Mol Biol Cell. 2005. PMID: 15537703 Free PMC article.
-
Fission yeast type 2 node proteins Blt1p and Gef2p cooperate to ensure timely completion of cytokinesis.BMC Mol Cell Biol. 2019 Jan 24;20(1):1. doi: 10.1186/s12860-018-0182-z. BMC Mol Cell Biol. 2019. PMID: 31041892 Free PMC article.
-
Proper actin ring formation and septum constriction requires coordinated regulation of SIN and MOR pathways through the germinal centre kinase MST-1.PLoS Genet. 2014 Apr 24;10(4):e1004306. doi: 10.1371/journal.pgen.1004306. eCollection 2014 Apr. PLoS Genet. 2014. PMID: 24762679 Free PMC article.
-
The multiprotein exocyst complex is essential for cell separation in Schizosaccharomyces pombe.Mol Biol Cell. 2002 Feb;13(2):515-29. doi: 10.1091/mbc.01-11-0542. Mol Biol Cell. 2002. PMID: 11854409 Free PMC article.
References
-
- Alfa C.E., Ducommun,B., Beach,D. and Hyams,J.S. (1990) Distinct nuclear and spindle pole body population of cyclin–cdc2 in fission yeast. Nature, 347, 680–682. - PubMed
-
- Balasubramanian M.K., McCollum,D. and Gould,K.L. (1997) Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol., 283, 494–506. - PubMed
-
- Barbet N., Muriel,W.J. and Carr,A.M. (1992) Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene, 114, 59–66. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

