Alteration of substrate and inhibitor specificity of feline immunodeficiency virus protease
- PMID: 10775609
- PMCID: PMC111993
- DOI: 10.1128/jvi.74.10.4710-4720.2000
Alteration of substrate and inhibitor specificity of feline immunodeficiency virus protease
Abstract
Feline immunodeficiency virus (FIV) protease is structurally very similar to human immunodeficiency virus (HIV) protease but exhibits distinct substrate and inhibitor specificities. We performed mutagenesis of subsite residues of FIV protease in order to define interactions that dictate this specificity. The I37V, N55M, M56I, V59I, and Q99V mutants yielded full activity. The I37V, N55M, V59I, and Q99V mutants showed a significant increase in activity against the HIV-1 reverse transcriptase/integrase and P2/nucleocapsid junction peptides compared with wild-type (wt) FIV protease. The I37V, V59I, and Q99V mutants also showed an increase in activity against two rapidly cleaved peptides selected by cleavage of a phage display library with HIV-1 protease. Mutations at Q54K, I98P, and L101I dramatically reduced activity. Mutants containing a I35D or I57G substitution showed no activity against either FIV or HIV substrates. FIV proteases all failed to cut HIV-1 matrix/capsid, P1/P6, P6/protease, and protease/reverse transcriptase junctions, indicating that none of the substitutions were sufficient to change the specificity completely. The I37V, N55M, M56I, V59I, and Q99V mutants, compared with wt FIV protease, all showed inhibitor specificity more similar to that of HIV-1 protease. The data also suggest that FIV protease prefers a hydrophobic P2/P2' residue like Val over Asn or Glu, which are utilized by HIV-1 protease, and that S2/S2' might play a critical role in distinguishing FIV and HIV-1 protease by specificity. The findings extend our observations regarding the interactions involved in substrate binding and aid in the development of broad-based inhibitors.
Figures

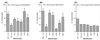
References
-
- Aiyar A, Leis J. Modification of the megaprimer method of PCR mutagenesis: improved amplification of the final product. Biotechniques. 1993;14:366–369. - PubMed
-
- Alewood P, Alewood D, Miranda L, Love S, Meutermans W, Wilson D. Rapid in situ neutralization protocols for Boc and Fmoc solid phase chemistries. Methods Enzymol. 1997;289:14–29. - PubMed
-
- Cameron C E, Ridky T W, Shulenin S, Leis J, Weber I T, Copeland T, Wlodawer A, Burstein H, Bizub-Bender D, Skalka A M. Mutational analysis of the substrate binding pockets of the Rous sarcoma virus and human immunodeficiency virus-1 proteases. J Biol Chem. 1994;269:11170–11177. - PubMed
-
- Dominy B N, Brooks C L., 3rd Methodology for protein-ligand binding studies: application to a model for drug resistance, the HIV/FIV protease system. Proteins. 1999;36:318–331. - PubMed
-
- Dunn B M, Pennington M W, Frase D C, Nash K. Comparison of inhibitor binding to feline and human immunodeficiency virus proteases: structure-based drug design and the resistance problem. Biopolymers. 1999;51:69–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

