Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses
- PMID: 10775610
- PMCID: PMC111994
- DOI: 10.1128/jvi.74.10.4721-4728.2000
Core protein phosphorylation modulates pregenomic RNA encapsidation to different extents in human and duck hepatitis B viruses
Abstract
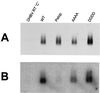
To clarify the role of core protein phosphorylation in pregenomic-RNA encapsidation of human and duck hepatitis B viruses (HBV and DHBV, respectively), we have examined the phosphorylation states of different forms of intracellular HBV core protein and the phenotypic effects of mutations in the phosphorylation sites of HBV and DHBV core proteins. We show that HBV core protein is phosphorylated to similar extents in the form of protein dimers and after further assembly in pregenomic RNA-containing capsids. Individual and multiple substitutions of alanine and aspartic acid for serine in the phosphorylation sites of HBV core protein resulted in site-specific and synergistic effects on RNA encapsidation, ranging from 2-fold enhancement to more than 10-fold inhibition. Core protein variants with mutations in all phosphorylation sites exhibited dominant-negative effects on RNA encapsidation by wild-type protein. The results suggest that the presence of phosphoserine at position 162 of HBV core protein is required for pregenomic-RNA encapsidation, whereas phosphoserine at position 170 optimizes the process and serine might be preferable in position 155. Examination of the pregenomic-RNA-encapsidating capacities of DHBV core protein variants, in which four phosphorylation sites were jointly mutated to alanine or aspartic acid, suggests that phosphorylation of DHBV core protein at these sites may optimize pregenomic-RNA encapsidation but that its impact is much less profound than in the case of HBV. The possible mechanisms by which RNA encapsidation may be modulated by core protein phosphorylation are discussed in the context of the observed differences between the two viruses.
Figures



Similar articles
-
Hepatitis B Virus Core Protein Dephosphorylation Occurs during Pregenomic RNA Encapsidation.J Virol. 2018 Jun 13;92(13):e02139-17. doi: 10.1128/JVI.02139-17. Print 2018 Jul 1. J Virol. 2018. PMID: 29669831 Free PMC article.
-
Capsid Phosphorylation State and Hepadnavirus Virion Secretion.J Virol. 2017 Apr 13;91(9):e00092-17. doi: 10.1128/JVI.00092-17. Print 2017 May 1. J Virol. 2017. PMID: 28228589 Free PMC article.
-
Chimeras of duck and heron hepatitis B viruses provide evidence for functional interactions between viral components of pregenomic RNA encapsidation.J Virol. 2004 Aug;78(16):8780-7. doi: 10.1128/JVI.78.16.8780-8787.2004. J Virol. 2004. PMID: 15280486 Free PMC article.
-
Duck hepatitis B virus (DHBV) as a model for understanding hepadnavirus neutralization.Arch Virol Suppl. 1993;8:133-9. doi: 10.1007/978-3-7091-9312-9_14. Arch Virol Suppl. 1993. PMID: 8260858 Review.
-
[The mechanisms of the translation of polymerase from HBV pregenomic RNA].Zhonghua Gan Zang Bing Za Zhi. 2021 Oct 20;29(10):1035-1040. doi: 10.3760/cma.j.cn501113-20210808-00384. Zhonghua Gan Zang Bing Za Zhi. 2021. PMID: 34814405 Review. Chinese.
Cited by
-
Importin β Can Bind Hepatitis B Virus Core Protein and Empty Core-Like Particles and Induce Structural Changes.PLoS Pathog. 2016 Aug 12;12(8):e1005802. doi: 10.1371/journal.ppat.1005802. eCollection 2016 Aug. PLoS Pathog. 2016. PMID: 27518410 Free PMC article.
-
Hepatitis B virus morphogenesis.World J Gastroenterol. 2007 Jan 7;13(1):65-73. doi: 10.3748/wjg.v13.i1.65. World J Gastroenterol. 2007. PMID: 17206755 Free PMC article. Review.
-
Core protein: A pleiotropic keystone in the HBV lifecycle.Antiviral Res. 2015 Sep;121:82-93. doi: 10.1016/j.antiviral.2015.06.020. Epub 2015 Jun 27. Antiviral Res. 2015. PMID: 26129969 Free PMC article. Review.
-
Cell-Free Hepatitis B Virus Capsid Assembly Dependent on the Core Protein C-Terminal Domain and Regulated by Phosphorylation.J Virol. 2016 May 27;90(12):5830-5844. doi: 10.1128/JVI.00394-16. Print 2016 Jun 15. J Virol. 2016. PMID: 27076641 Free PMC article.
-
Cyclin-dependent kinase 2 phosphorylates s/t-p sites in the hepadnavirus core protein C-terminal domain and is incorporated into viral capsids.J Virol. 2012 Nov;86(22):12237-50. doi: 10.1128/JVI.01218-12. Epub 2012 Sep 5. J Virol. 2012. PMID: 22951823 Free PMC article.
References
-
- Beames B, Lanford R E. Carboxy-terminal truncations of the HBV core protein affect capsid formation and the apparent size of encapsidated HBV RNA. Virology. 1993;194:597–607. - PubMed
-
- Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. - PubMed
-
- Ganem D. Hepadnaviridae: the viruses and their replication. In: Fields B, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Raven Publishers; 1996. pp. 2703–2737.
-
- Gorman C. High efficiency gene transfer into mammalian cells. In: Glover D M, editor. DNA cloning: a practical approach. Oxford, United Kingdom: IRL Press; 1985. pp. 143–190.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

