Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells
- PMID: 10775618
- PMCID: PMC112002
- DOI: 10.1128/jvi.74.10.4795-4806.2000
Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells
Abstract
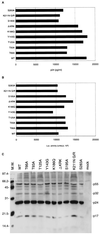
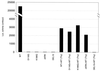
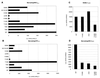
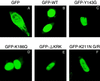
Human immunodeficiency virus type 1 integrase (HIV-1 IN) is thought to have several putative roles at steps prior to integration, such as reverse transcription and nuclear transport of the preintegration complex (PIC). Here, we investigated new functional aspects of HIV-1 IN in the context of the viral replication cycle through point mutagenesis of Ser, Thr, Tyr, Lys, and Arg residues conserved in IN, some of which are located at possible phosphorylation sites. Our results showed that mutations of these Ser or Thr residues had no effect on reverse transcription and nuclear transport of PIC but had a slight effect on integration. Of note, mutations in the conserved KRK motif (amino acids 186 to 189), proposed previously as a putative nuclear localization signal (NLS) of HIV-1 IN, did not affect the karyophilic property of HIV-1 IN as shown by using a green fluorescent protein fusion protein expression system. Instead, these KRK mutations resulted in an almost complete lack of viral gene expression due to the failure to complete reverse transcription. This defect was complemented by supplying wild-type IN in trans, suggesting a trans-acting function of the KRK motif of IN in reverse transcription. Mutation at the conserved Tyr 143 (Y143G) resulted in partial impairment of completion of reverse transcription in monocyte-derived macrophages (MDM) but not in rhabdomyosarcoma cells. Similar effects were obtained by introducing a stop codon in the vpr gene (DeltaVpr), and additive effects of both mutations (Y143G plus DeltaVpr) were observed. In addition, these mutants did not produce two-long terminal repeat DNA, a surrogate marker for nuclear entry, in MDM. Thus, the possible impairment of Y143G might occur during the nuclear transport of the PIC. Taken together, our results identified new functional aspects of the conserved residues in HIV-1 IN: i) the KRK motif might have a role in efficient reverse transcription in both dividing and nondividing cells but not in the NLS function; ii) Y143 might be an important residue for maintaining efficient proviral DNA formation in nondividing cells.
Figures




Similar articles
-
Contribution of the C-terminal tri-lysine regions of human immunodeficiency virus type 1 integrase for efficient reverse transcription and viral DNA nuclear import.Retrovirology. 2005 Oct 18;2:62. doi: 10.1186/1742-4690-2-62. Retrovirology. 2005. PMID: 16232319 Free PMC article.
-
The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA.J Virol. 2000 Aug;74(15):7119-26. doi: 10.1128/jvi.74.15.7119-7126.2000. J Virol. 2000. PMID: 10888652 Free PMC article.
-
Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex.J Mol Biol. 2000 Jun 2;299(2):359-68. doi: 10.1006/jmbi.2000.3768. J Mol Biol. 2000. PMID: 10860744
-
Nuclear import of the pre-integration complex (PIC): the Achilles heel of HIV?Curr Drug Targets. 2003 Jul;4(5):409-29. doi: 10.2174/1389450033490984. Curr Drug Targets. 2003. PMID: 12816349 Review.
-
Viral protein R of HIV-1.Rev Med Virol. 1999 Jan-Mar;9(1):39-49. doi: 10.1002/(sici)1099-1654(199901/03)9:1<39::aid-rmv235>3.0.co;2-3. Rev Med Virol. 1999. PMID: 10371671 Review.
Cited by
-
Correlation of recombinant integrase activity and functional preintegration complex formation during acute infection by replication-defective integrase mutant human immunodeficiency virus.J Virol. 2012 Apr;86(7):3861-79. doi: 10.1128/JVI.06386-11. Epub 2012 Jan 25. J Virol. 2012. PMID: 22278243 Free PMC article.
-
Quantitative prediction of integrase inhibitor resistance from genotype through consensus linear regression modeling.Virol J. 2013 Jan 3;10:8. doi: 10.1186/1743-422X-10-8. Virol J. 2013. PMID: 23282253 Free PMC article.
-
The cell cycle independence of HIV infections is not determined by known karyophilic viral elements.PLoS Pathog. 2005 Nov;1(3):e18. doi: 10.1371/journal.ppat.0010018. Epub 2005 Nov 11. PLoS Pathog. 2005. PMID: 16292356 Free PMC article.
-
Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication.J Virol. 2004 Dec;78(23):12735-46. doi: 10.1128/JVI.78.23.12735-12746.2004. J Virol. 2004. PMID: 15542626 Free PMC article.
-
Molecular dynamics studies of the wild-type and double mutant HIV-1 integrase complexed with the 5CITEP inhibitor: mechanism for inhibition and drug resistance.Biophys J. 2003 Mar;84(3):1450-63. doi: 10.1016/S0006-3495(03)74958-3. Biophys J. 2003. PMID: 12609852 Free PMC article.
References
-
- Balliet J W, Kolson D L, Eiger G, Kim F M, McGann K A, Srinivasan A, Collman R. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology. 1994;200:623–631. - PubMed
-
- Bukrinsky M I, Haffar O K. HIV-1 nuclear import: in search of a leader. Front Biosci. 1997;2:578–587. - PubMed
-
- Burke C J, Sanyal G, Bruner M W, Ryan J A, LaFemina R L, Robbins H L, Zeft A S, Middaugh C R, Cordingley M G. Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J Biol Chem. 1992;267:9639–9644. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

