A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene
- PMID: 10775623
- PMCID: PMC112007
- DOI: 10.1128/jvi.74.10.4839-4852.2000
A Rev-independent human immunodeficiency virus type 1 (HIV-1)-based vector that exploits a codon-optimized HIV-1 gag-pol gene
Abstract
The human immunodeficiency virus (HIV) genome is AU rich, and this imparts a codon bias that is quite different from the one used by human genes. The codon usage is particularly marked for the gag, pol, and env genes. Interestingly, the expression of these genes is dependent on the presence of the Rev/Rev-responsive element (RRE) regulatory system, even in contexts other than the HIV genome. The Rev dependency has been explained in part by the presence of RNA instability sequences residing in these coding regions. The requirement for Rev also places a limitation on the development of HIV-based vectors, because of the requirement to provide an accessory factor. We have now synthesized a complete codon-optimized HIV-1 gag-pol gene. We show that expression levels are high and that expression is Rev independent. This effect is due to an increase in the amount of gag-pol mRNA. Provision of the RRE in cis did not lower protein or RNA levels or stimulate a Rev response. Furthermore we have used this synthetic gag-pol gene to produce HIV vectors that now lack all of the accessory proteins. These vectors should now be safer than murine leukemia virus-based vectors.
Figures







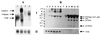

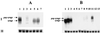



Similar articles
-
Helper plasmids for production of HIV-1-derived vectors.Hum Gene Ther. 2001 Nov 20;12(17):2081-93. doi: 10.1089/10430340152677421. Hum Gene Ther. 2001. PMID: 11747598
-
Rev-independent expression of synthetic gag-pol genes of human immunodeficiency virus type 1 and simian immunodeficiency virus: implications for the safety of lentiviral vectors.Hum Gene Ther. 2000 Nov 20;11(17):2403-13. doi: 10.1089/104303400750038507. Hum Gene Ther. 2000. PMID: 11096444
-
Interactions of INS (CRS) elements and the splicing machinery regulate the production of Rev-responsive mRNAs.J Mol Biol. 1996 Mar 29;257(2):246-64. doi: 10.1006/jmbi.1996.0160. J Mol Biol. 1996. PMID: 8609621
-
The origin of human immunodeficiency virus type-1 rev gene. An evolutionary hypothesis.FEBS Lett. 1994 Jan 31;338(2):118-21. doi: 10.1016/0014-5793(94)80347-1. FEBS Lett. 1994. PMID: 8307167 Review.
-
Gene delivery by lentivirus vectors an overview.Methods Mol Biol. 2004;246:367-90. doi: 10.1385/1-59259-650-9:367. Methods Mol Biol. 2004. PMID: 14970605 Review.
Cited by
-
Capsid-Targeted Viral Inactivation: A Novel Tactic for Inhibiting Replication in Viral Infections.Viruses. 2016 Sep 21;8(9):258. doi: 10.3390/v8090258. Viruses. 2016. PMID: 27657114 Free PMC article. Review.
-
Transfection of nerve cells.Neurosci Behav Physiol. 2010 Mar;40(3):269-77. doi: 10.1007/s11055-010-9254-z. Epub 2010 Feb 10. Neurosci Behav Physiol. 2010. PMID: 20146014
-
Titers of lentiviral vectors encoding shRNAs and miRNAs are reduced by different mechanisms that require distinct repair strategies.RNA. 2010 Jul;16(7):1328-39. doi: 10.1261/rna.1887910. Epub 2010 May 24. RNA. 2010. PMID: 20498457 Free PMC article.
-
Design of a novel integration-deficient lentivector technology that incorporates genetic and posttranslational elements to target human dendritic cells.Mol Ther. 2014 Mar;22(3):575-587. doi: 10.1038/mt.2013.278. Epub 2013 Dec 6. Mol Ther. 2014. PMID: 24419083 Free PMC article.
-
Multiple barriers to recombination between divergent HIV-1 variants revealed by a dual-marker recombination assay.J Mol Biol. 2011 Apr 8;407(4):521-31. doi: 10.1016/j.jmb.2011.01.052. Epub 2011 Feb 3. J Mol Biol. 2011. PMID: 21295586 Free PMC article.
References
-
- Afonina E, Neumann M, Pavlakis G. Preferential binding of poly(A)-binding protein I to an inhibitory RNA element in the HIV-1 gag mRNA. J Biol Chem. 1997;272:2307–2311. - PubMed
-
- Arai T, Takada M, Ui M, Iba H. Dose-dependent transduction of vesicular stomatitis virus G protein-pseudotyped retrovirus vector into human solid tumor cell lines and murine fibroblasts. Virology. 1999;260:109–115. - PubMed
-
- Arrigo S J, Chen I S. Rev is necessary for translation but not cytoplasmic accumulation of HIV-1 vif, vpr, and env/vpu 2 RNAs. Genes Dev. 1991;5:808–819. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

