Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus
- PMID: 10775636
- PMCID: PMC112020
- DOI: 10.1128/jvi.74.10.4919-4928.2000
Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus
Abstract
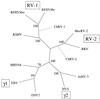
We have cloned and characterized the entire DNA polymerase gene and flanking regions from Kaposi's sarcoma-associated herpesvirus (KSHV) and two closely related macaque homologs of KSHV, retroperitoneal fibromatosis-associated herpesvirus-Macaca nemestrina (RFHVMn) and -Macaca mulatta (RFHVMm). We have also identified and partially characterized the corresponding genomic region of another KSHV-like herpesvirus, provisionally named "M. nemestrina rhadinovirus type 2 (MneRV-2)," with close similarity to rhesus rhadinovirus (RRV). A sequence comparison of these four macaque viruses and two KSHV-like gammaherpesviruses recently identified in African green monkeys, Chlorocebus rhadinovirus types 1 and 2 (ChRV-1 and ChRV-2) reveals the presence of two distinct lineages of KSHV-like rhadinoviruses in Old World primates. The first rhadinovirus lineage consists of KSHV and its closely related homologs RFHVMn, RFHVMm, and ChRV-1, while the second more distantly related lineage consists of RRV, MneRV-2, and ChRV-2. Our findings raise the possibility of the existence of another human KSHV-like herpesvirus belonging to the second rhadinovirus lineage.
Figures





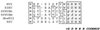
References
-
- Chang Y, Moore P S. Kaposi's sarcoma (KS)-associated herpesvirus and its role in KS. Infect Agents Dis. 1996;5:215–222. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

