DNA methyltransferase contributes to delayed ischemic brain injury
- PMID: 10777781
- PMCID: PMC6773114
- DOI: 10.1523/JNEUROSCI.20-09-03175.2000
DNA methyltransferase contributes to delayed ischemic brain injury
Abstract
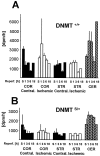
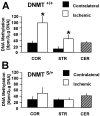
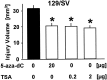
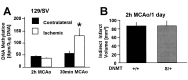
DNA methylation is important for controlling the profile of gene expression and is catalyzed by DNA methyltransferase (MTase), an enzyme that is abundant in brain. Because significant DNA damage and alterations in gene expression develop as a consequence of cerebral ischemia, we measured MTase activity in vitro and DNA methylation in vivo after mild focal brain ischemia. After 30 min middle cerebral artery occlusion (MCAo) and reperfusion, MTase catalytic activity and the 190 kDa band on immunoblot did not change over time. However, [(3)H]methyl-group incorporation into DNA increased significantly in wild-type mice after reperfusion, but not in mutant mice heterozygous for a DNA methyltransferase gene deletion (Dnmt(S/+)). Dnmt(S/+) mice were resistant to mild ischemic damage, suggesting that increased DNA methylation is associated with augmented brain injury after MCA occlusion. Consistent with this formulation, treatment with the MTase inhibitor 5-aza-2'-deoxycytidine and the deacetylation inhibitor trichostatin A conferred stroke protection in wild-type mice. In contrast to mild stroke, however, DNA methylation was not enhanced, and reduced dnmt gene expression was not protective in an ischemia model of excitotoxic/necrotic cell death. In conclusion, our results demonstrate that MTase activity contributes to poor tissue outcome after mild ischemic brain injury.
Figures

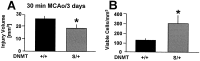
Similar articles
-
Relationship between metabolic dysfunctions, gene responses and delayed cell death after mild focal cerebral ischemia in mice.Neuroscience. 2001;104(4):947-55. doi: 10.1016/s0306-4522(01)00125-7. Neuroscience. 2001. PMID: 11457582
-
Resveratrol attenuates ischemic brain damage in the delayed phase after stroke and induces messenger RNA and protein express for angiogenic factors.J Vasc Surg. 2008 Sep;48(3):709-14. doi: 10.1016/j.jvs.2008.04.007. Epub 2008 Jun 24. J Vasc Surg. 2008. PMID: 18572362
-
Effects of cerebral ischemia in mice lacking DNA methyltransferase 1 in post-mitotic neurons.Neuroreport. 2001 Dec 4;12(17):3763-6. doi: 10.1097/00001756-200112040-00032. Neuroreport. 2001. PMID: 11726790
-
Coicis semen protects against focal cerebral ischemia-reperfusion injury by inhibiting oxidative stress and promoting angiogenesis via the TGFβ/ALK1/Smad1/5 signaling pathway.Aging (Albany NY). 2020 Nov 16;13(1):877-893. doi: 10.18632/aging.202194. Epub 2020 Nov 16. Aging (Albany NY). 2020. PMID: 33290255 Free PMC article.
-
Downregulation of Nogo-B ameliorates cerebral ischemia/reperfusion injury in mice through regulating microglia polarization via TLR4/NF-kappaB pathway.Neurochem Int. 2023 Jul;167:105553. doi: 10.1016/j.neuint.2023.105553. Epub 2023 May 23. Neurochem Int. 2023. PMID: 37230196 Review.
Cited by
-
Differential DNA methylation profiles of coding and non-coding genes define hippocampal sclerosis in human temporal lobe epilepsy.Brain. 2015 Mar;138(Pt 3):616-31. doi: 10.1093/brain/awu373. Epub 2014 Dec 30. Brain. 2015. PMID: 25552301 Free PMC article.
-
Tolerance to ischemia - an increasingly complex biology.Transl Stroke Res. 2013 Feb;4(1):40-50. doi: 10.1007/s12975-012-0246-x. Epub 2013 Jan 11. Transl Stroke Res. 2013. PMID: 23504451 Free PMC article. Review.
-
Differential roles of epigenetic regulators in the survival and differentiation of oligodendrocyte precursor cells.Glia. 2019 Apr;67(4):718-728. doi: 10.1002/glia.23567. Epub 2018 Nov 28. Glia. 2019. PMID: 30793389 Free PMC article.
-
Stroke Causes DNA Methylation at Ncx1 Heart Promoter in the Brain Via DNMT1/MeCP2/REST Epigenetic Complex.J Am Heart Assoc. 2024 Mar 19;13(6):e030460. doi: 10.1161/JAHA.123.030460. Epub 2024 Mar 8. J Am Heart Assoc. 2024. PMID: 38456444 Free PMC article.
-
Epigenetics in Stroke Recovery.Genes (Basel). 2017 Feb 27;8(3):89. doi: 10.3390/genes8030089. Genes (Basel). 2017. PMID: 28264471 Free PMC article. Review.
References
-
- Adams RL, Burdon RH. DNA methylation in eukaryotes. CRC Crit Rev Biochem. 1982;13:349–384. - PubMed
-
- Baylin SB, Makos M, Wu JJ, Yen RW, de Bustros A, Vertino P, Nelkin BD. Abnormal patterns of DNA methylation in human neoplasia: potential consequences for tumor progression. Cancer Cells. 1991;3:383–390. - PubMed
-
- Bestor TH. DNA methylation: evolution of a bacterial immune function into a regulator of gene expression and genome structure in higher eukaryotes. Philos Trans R Soc Lond B Biol Sci. 1990;326:179–187. - PubMed
-
- Bhattacharya SK, Rachmandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. - PubMed
-
- Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
