SINE retroposons can be used in vivo as nucleation centers for de novo methylation
- PMID: 10779333
- PMCID: PMC85636
- DOI: 10.1128/MCB.20.10.3434-3441.2000
SINE retroposons can be used in vivo as nucleation centers for de novo methylation
Abstract
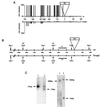
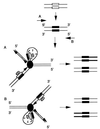
SINEs (short interspersed elements) are an abundant class of transposable elements found in a wide variety of eukaryotes. Using the genomic sequencing technique, we observed that plant S1 SINE retroposons mainly integrate in hypomethylated DNA regions and are targeted by methylases. Methylation can then spread from the SINE into flanking genomic sequences, creating distal epigenetic modifications. This methylation spreading is vectorially directed upstream or downstream of the S1 element, suggesting that it could be facilitated when a potentially good methylatable sequence is single stranded during DNA replication, particularly when located on the lagging strand. Replication of a short methylated DNA region could thus lead to the de novo methylation of upstream or downstream adjacent sequences.
Figures


References
-
- Bennetzen J L. The mutator transposable element system of maize. Curr Top Microbiol Immunol. 1996;24:195–229. - PubMed
-
- Bird A. The essential of DNA methylation. Cell. 1992;70:5–8. - PubMed
-
- Carotti D, Funiciello S, Palitti F, Strom R. Influence of pre-existing methylation on the de novo activity of eukaryotic DNA methyltransferase. Biochemistry. 1998;37:1101–1108. - PubMed
-
- Chesnokov I, Schmid C W. Flanking sequences of an Alu source stimulate transcription in vitro by interacting with sequence-specific transcription factors. J Mol Evol. 1996;42:30–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
