Binding of equine infectious anemia virus rev to an exon splicing enhancer mediates alternative splicing and nuclear export of viral mRNAs
- PMID: 10779344
- PMCID: PMC85647
- DOI: 10.1128/MCB.20.10.3550-3557.2000
Binding of equine infectious anemia virus rev to an exon splicing enhancer mediates alternative splicing and nuclear export of viral mRNAs
Abstract
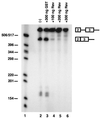
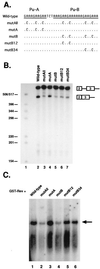
In addition to facilitating the nuclear export of incompletely spliced viral mRNAs, equine infectious anemia virus (EIAV) Rev regulates alternative splicing of the third exon of the tat/rev mRNA. In the presence of Rev, this exon of the bicistronic RNA is skipped in a fraction of the spliced mRNAs. In this report, the cis-acting requirements for exon 3 usage were correlated with sequences necessary for Rev binding and transport of incompletely spliced RNA. The presence of a purine-rich exon splicing enhancer (ESE) was required for exon 3 recognition, and the addition of Rev inhibited exon 3 splicing. Glutathione-S-transferase (GST)-Rev bound to probes containing the ESE, and mutation of GAA repeats to GCA within the ESE inhibited both exon 3 recognition in RNA splicing experiments and GST-Rev binding in vitro. These results suggest that Rev regulates alternative splicing by binding at or near the ESE to block SR protein-ESE interactions. A 57-nucleotide sequence containing the ESE was sufficient to mediate Rev-dependent nuclear export of incompletely spliced RNAs. Rev export activity was significantly inhibited by mutation of the ESE or by trans-complementation with SF2/ASF. These results indicate that the ESE functions as a Rev-responsive element and demonstrate that EIAV Rev mediates exon 3 exclusion through protein-RNA interactions required for efficient export of incompletely spliced viral RNAs.
Figures





References
-
- Ali S A, Steinkasserer A. PCR-ligation-PCR mutagenesis: a protocol for creating gene fusions and mutations. Biotechniques. 1995;18:746–750. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
