The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae
- PMID: 10779558
- PMCID: PMC25801
- DOI: 10.1073/pnas.090102597
The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1-methyladenosine) methyltransferase of Saccharomyces cerevisiae
Abstract
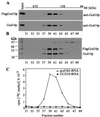
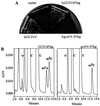
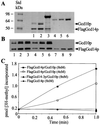
The modified nucleoside 1-methyladenosine (m(1)A) is found at position 58 in the TPsiC loop of many eukaryotic tRNAs. The absence of m(1)A from all tRNAs in Saccharomyces cerevisiae mutants lacking Gcd10p elicits severe defects in processing and stability of initiator methionine tRNA (tRNA(i)(Met)). Gcd10p is found in a complex with Gcd14p, which contains conserved motifs for binding S-adenosylmethionine (AdoMet). These facts, plus our demonstration that gcd14Delta cells lacked m(1)A, strongly suggested that Gcd10p/Gcd14p complex is the yeast tRNA(m(1)A)methyltransferase [(m(1)A)MTase]. Supporting this prediction, affinity-purified Gcd10p/Gcd14p complexes used AdoMet as a methyl donor to synthesize m(1)A in either total tRNA or purified tRNA(i)(Met) lacking only this modification. Kinetic analysis of the purified complex revealed K(M) values for AdoMet or tRNA(i)(Met) of 5.0 microM and 2.5 nM, respectively. Mutations in the predicted AdoMet-binding domain destroyed GCD14 function in vivo and (m(1)A)MTase activity in vitro. Purified Flag-tagged Gcd14p alone had no enzymatic activity and was severely impaired for tRNA-binding compared with the wild-type complex, suggesting that Gcd10p is required for tight binding of the tRNA substrate. Our results provide a demonstration of a two-component tRNA MTase and suggest that binding of AdoMet and tRNA substrates depends on different subunits of the complex.
Figures

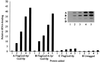
References
-
- Björk G R. In: tRNA: Structure Biosynthesis and Function. Söll D, RajBhandary U L, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 165–206.
-
- Björk G R, Ericson J U, Gustafsson C E, Hagervall T G, Jönsson Y H, Wikström P M. Annu Rev Biochem. 1987;56:263–287. - PubMed
-
- Åström S U, Byström A S. Cell. 1994;79:535–546. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

