The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens
- PMID: 10781104
- PMCID: PMC18339
- DOI: 10.1073/pnas.97.9.4956
The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens
Abstract
An integrated molecular and physiological investigation of the fundamental mechanisms of heavy metal accumulation was conducted in Thlaspi caerulescens, a Zn/Cd-hyperaccumulating plant species. A heavy metal transporter cDNA, ZNT1, was cloned from T. caerulescens through functional complementation in yeast and was shown to mediate high-affinity Zn(2+) uptake as well as low-affinity Cd(2+) uptake. It was found that this transporter is expressed at very high levels in roots and shoots of the hyperaccumulator. A study of ZNT1 expression and high-affinity Zn(2+) uptake in roots of T. caerulescens and in a related nonaccumulator, Thlaspi arvense, showed that alteration in the regulation of ZNT1 gene expression by plant Zn status results in the overexpression of this transporter and in increased Zn influx in roots of the hyperaccumulating Thlaspi species. These findings yield insights into the molecular regulation and control of plant heavy metal and micronutrient accumulation and homeostasis, as well as provide information that will contribute to the advancement of phytoremediation by the future engineering of plants with improved heavy metal uptake and tolerance.
Figures


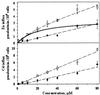


References
-
- Salt D E, Blaylock M, Kumar N P B A, Dushenkov V, Ensley B D, Chet I, Raskin I. Bio/Technology. 1995;13:468–474. - PubMed
-
- Salt D E, Smith R D, Raskin I. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:643–668. - PubMed
-
- Baker A J M. J Plant Nutr. 1981;3:643–654.
-
- Baker A J M, Brooks R R. Biorecovery. 1989;1:81–126.
-
- Brown S L, Chaney R L, Angle J S, Baker A J M. Soil Sci Soc Am J. 1995;59:125–132.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

