Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases
- PMID: 10781544
- PMCID: PMC101984
- DOI: 10.1128/JB.182.10.2761-2770.2000
Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases
Abstract
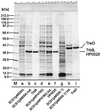
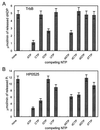
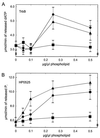
Type IV secretion systems direct transport of protein or nucleoprotein complexes across the cell envelopes of prokaryotic donor and eukaryotic or prokaryotic recipient cells. The process is mediated by a membrane-spanning multiprotein assembly. Potential NTPases belonging to the VirB11 family are an essential part of the membrane-spanning complex. Three representatives of these NTPases originating from the conjugative transfer regions of plasmids RP4 (TrbB) and R388 (TrwD) and from the cag pathogenicity island of Helicobacter pylori (HP0525) were overproduced and purified in native form. The proteins display NTPase activity with distinct substrate specificities in vitro. TrbB shows its highest specific hydrolase activity with dATP, and the preferred substrate for HP0525 is ATP. Analysis of defined TrbB mutations altered in motifs conserved within the VirB11 protein family shows that there is a correlation between the loss or reduction of NTPase activity and transfer frequency. Tryptophan fluorescence spectroscopy of TrbB and HP0525 suggests that both interact with phospholipid membranes, changing their conformation. NTPase activity of both proteins was stimulated by the addition of certain phospholipids. According to our results, Virb11-like proteins seem to most likely be involved in the assembly of the membrane-spanning multiprotein complex.
Figures



References
-
- Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Pontén T, Alsmark U C M, Podowski R M, Kurland C G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. - PubMed
-
- Binet R, Letoffe S, Ghigo J M, Delepelaire P, Wandersman C. Protein secretion by Gram-negative bacterial ABC exporters—a review. Gene. 1997;192:7–11. - PubMed
-
- Bordo D, Argos P. Suggestions for “safe” residue substitutions in site-directed mutageneses. J Mol Biol. 1991;217:721–729. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

