Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins
- PMID: 10783164
- PMCID: PMC316538
Mcm10 and the MCM2-7 complex interact to initiate DNA synthesis and to release replication factors from origins
Abstract
MCM2-7, a complex of six subunits, is an essential component of the prereplication chromatin that is assembled at Saccharomyces cerevisiae replication origins during G(1) phase. It is also believed to be the processive helicase at growing forks. To elucidate the action of MCM2-7 during the transition from initiation to elongation replication, we have focused our studies on Mcm10, a replication initiation protein that physically interacts with members of the MCM2-7 complex. We show that Mcm10 is a chromatin-associated protein that mediates the association of the MCM2-7 complex with replication origins. Furthermore, diminished interaction between Mcm10 and Mcm7, a subunit of the MCM2-7 complex, by a mutation in either Mcm10 or Mcm7 inhibits replication initiation. Surprisingly, a double mutant containing both the mcm10-1 and mcm7-1 (cdc47-1) alleles restores interaction between Mcm10 and Mcm7 and corrects all of the defects exhibited by each of the single mutants, including the stalling of replication forks at replication origins typically seen in mcm10-1 cells. This mutual compensation of defects between two independently isolated mutations is allele specific. These results suggest that Mcm10, like Mcm7, is a critical component of the prereplication chromatin and that interaction between Mcm10 and Mcm7 is required for proper replication initiation and prompt release of origin-bound factors.
Figures
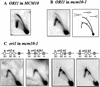

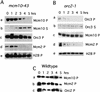







Similar articles
-
A lesion in the DNA replication initiation factor Mcm10 induces pausing of elongation forks through chromosomal replication origins in Saccharomyces cerevisiae.Mol Cell Biol. 1997 Jun;17(6):3261-71. doi: 10.1128/MCB.17.6.3261. Mol Cell Biol. 1997. PMID: 9154825 Free PMC article.
-
Budding yeast mcm10/dna43 mutant requires a novel repair pathway for viability.Genes Cells. 2003 May;8(5):465-80. doi: 10.1046/j.1365-2443.2003.00648.x. Genes Cells. 2003. PMID: 12694535
-
Interactions between Mcm10p and other replication factors are required for proper initiation and elongation of chromosomal DNA replication in Saccharomyces cerevisiae.Genes Cells. 2000 Dec;5(12):975-89. doi: 10.1046/j.1365-2443.2000.00387.x. Genes Cells. 2000. PMID: 11168584
-
Initiating DNA synthesis: from recruiting to activating the MCM complex.J Cell Sci. 2001 Apr;114(Pt 8):1447-54. doi: 10.1242/jcs.114.8.1447. J Cell Sci. 2001. PMID: 11282021 Review.
-
Getting a grip on licensing: mechanism of stable Mcm2-7 loading onto replication origins.Mol Cell. 2006 Jan 20;21(2):143-4. doi: 10.1016/j.molcel.2006.01.003. Mol Cell. 2006. PMID: 16427002 Review.
Cited by
-
Stepwise assembly of initiation proteins at budding yeast replication origins in vitro.Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14115-20. doi: 10.1073/pnas.97.26.14115. Proc Natl Acad Sci U S A. 2000. PMID: 11121019 Free PMC article.
-
Common Trajectories of Highly Effective CD19-Specific CAR T Cells Identified by Endogenous T-cell Receptor Lineages.Cancer Discov. 2022 Sep 2;12(9):2098-2119. doi: 10.1158/2159-8290.CD-21-1508. Cancer Discov. 2022. PMID: 35792801 Free PMC article.
-
MCM10: one tool for all-Integrity, maintenance and damage control.Semin Cell Dev Biol. 2014 Jun;30:121-30. doi: 10.1016/j.semcdb.2014.03.017. Epub 2014 Mar 21. Semin Cell Dev Biol. 2014. PMID: 24662891 Free PMC article. Review.
-
Hexameric ring structure of human MCM10 DNA replication factor.EMBO Rep. 2007 Oct;8(10):925-30. doi: 10.1038/sj.embor.7401064. Epub 2007 Sep 7. EMBO Rep. 2007. PMID: 17823614 Free PMC article.
-
Distinct genomic aberrations associated with ERG rearranged prostate cancer.Genes Chromosomes Cancer. 2009 Apr;48(4):366-80. doi: 10.1002/gcc.20647. Genes Chromosomes Cancer. 2009. PMID: 19156837 Free PMC article.
References
-
- Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. - PubMed
-
- Ausubel FM, Brent T, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. Harvard, MA: John Wiley & Sons; 1998.
-
- Aves S, Tongue N, Foster AJ. The essential Schizosaccharomyces pombe cdc23 DNA replication gene shares structural and functional homology with the Saccharomyces cerevisiae DNA43 (MCM10) gene. Curr Genet. 1998;34:164–171. - PubMed
-
- Bell S, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
