Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum
- PMID: 10783170
- PMCID: PMC316543
Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum
Abstract
Medulloblastomas are among the most common malignancies in childhood, and they are associated with substantial mortality and morbidity. The molecular pathogenesis as well as the ontogeny of these neoplasms is still poorly understood. We have generated a mouse model for medulloblastoma by Cre-LoxP-mediated inactivation of Rb and p53 tumor suppressor genes in the cerebellar external granular layer (EGL) cells. GFAP-Cre-mediated recombination was found both in astrocytes and in immature precursor cells of the EGL in the developing cerebellum. GFAP-Cre;Rb(LoxP/LoxP);p53(-/- or LoxP/LoxP) mice developed highly aggressive embryonal tumors of the cerebellum with typical features of medulloblastoma. These tumors were identified as early as 7 weeks of age on the outer surface of the molecular layer, corresponding to the location of the EGL cells during development. Our results demonstrate that loss of function of RB is essential for medulloblastoma development in the mouse and strongly support the hypothesis that medulloblastomas arise from multipotent precursor cells located in the EGL.
Figures








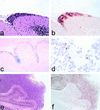
References
-
- Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kageyama R. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8738. - PubMed
-
- Badiali M, Pession A, Basso G, Andreini L, Rigobello L, Galassi E, Giangaspero F. N-myc and c-myc oncogenes amplification in medulloblastomas. Evidence of particularly aggressive behavior of a tumor with c-myc amplification. Tumori. 1991;77:118–121. - PubMed
-
- Bailey P, Cushing H. Medulloblastoma cerebelli: Common type midcerebellar glioma of childhood. Arch Neurol Psych. 1925;14:192–223.
-
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
