A solid-state NMR index of helical membrane protein structure and topology
- PMID: 10783285
- PMCID: PMC3437921
- DOI: 10.1006/jmre.2000.2035
A solid-state NMR index of helical membrane protein structure and topology
Abstract
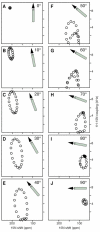
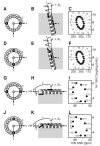
The secondary structure and topology of membrane proteins can be described by inspection of two-dimensional (1)H-(15)N dipolar coupling/(15)N chemical shift polarization inversion spin exchange at the magic angle spectra obtained from uniformly (15)N-labeled samples in oriented bilayers. The characteristic wheel-like patterns of resonances observed in these spectra reflect helical wheel projections of residues in both transmembrane and in-plane helices and hence provide direct indices of the secondary structure and topology of membrane proteins in phospholipid bilayers. We refer to these patterns as PISA (polarity index slant angle) wheels. The transmembrane helix of the M2 peptide corresponding to the pore-lining segment of the acetylcholine receptor and the membrane surface helix of the antibiotic peptide magainin are used as examples.
Copyright 2000 Academic Press.
Figures

References
-
- Opella SJ. NMR and membrane proteins. Nat. Struct. Biol. NMR I Suppl. 1997;4:845–848. - PubMed
-
- Griffin R. Dipolar recoupling in MAS spectra of biological solids. Nat. Struct. Biol. NMR II Suppl. 1998;5:508–512. - PubMed
-
- Smith SO, Ascheim K, Groesbeck M. Magic angle spinning NMR spectroscopy of membrane proteins. Q. Rev. Biophys. 1996;29:395–449. - PubMed
-
- McDowell LM, Schaefer J. High resolution NMR of biological solids. Curr. Opin. Struct. Biol. 1996;6:624–629. - PubMed
-
- Glaubitz C, Watts A. Magic angle-oriented sample spinning (MAOSS): A new approach toward biomembrane studies. J. Magn. Reson. 1998;130:305–316. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

