Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract
- PMID: 10788396
- PMCID: PMC101469
- DOI: 10.1128/AEM.66.5.2166-2174.2000
Molecular ecological analysis of the succession and diversity of sulfate-reducing bacteria in the mouse gastrointestinal tract
Abstract
Intestinal sulfate-reducing bacteria (SRB) growth and resultant hydrogen sulfide production may damage the gastrointestinal epithelium and thereby contribute to chronic intestinal disorders. However, the ecology and phylogenetic diversity of intestinal dissimilatory SRB populations are poorly understood, and endogenous or exogenous sources of available sulfate are not well defined. The succession of intestinal SRB was therefore compared in inbred C57BL/6J mice using a PCR-based metabolic molecular ecology (MME) approach that targets a conserved region of subunit A of the adenosine-5'-phosphosulfate (APS) reductase gene. The APS reductase-based MME strategy revealed intestinal SRB in the stomach and small intestine of 1-, 4-, and 7-day-old mice and throughout the gastrointestinal tract of 14-, 21-, 30-, 60-, and 90-day-old mice. Phylogenetic analysis of APS reductase amplicons obtained from the stomach, middle small intestine, and cecum of neonatal mice revealed that Desulfotomaculum spp. may be a predominant SRB group in the neonatal mouse intestine. Dot blot hybridizations with SRB-specific 16S ribosomal DNA (rDNA) probes demonstrated SRB colonization of the cecum and colon pre- and postweaning and colonization of the stomach and small intestine of mature mice only. The 16S rDNA hybridization data further demonstrated that SRB populations were most numerous in intestinal regions harboring sulfomucin-containing goblet cells, regardless of age. Reverse transcriptase PCR analysis demonstrated APS reductase mRNA expression in all intestinal segments of 30-day-old mice, including the stomach. These results demonstrate for the first time widespread colonization of the mouse intestine by dissimilatory SRB and evidence of spatial-specific SRB populations and sulfomucin patterns along the gastrointestinal tract.
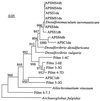
Figures






References
-
- Beerens H, Romond C. Sulfate-reducing anaerobic bacteria in human feces. Am J Clin Nutr. 1977;30:1770–1776. - PubMed
-
- Corfield A P, Wagner S A, Clamp J R, Kriaris M S, Hoskins L C. Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase and glycosulfatase activities by strains of faecal bacteria. Infect Immun. 1992;66:3971–3978. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

