Molecular differentiation of seven Malassezia species
- PMID: 10790115
- PMCID: PMC86611
- DOI: 10.1128/JCM.38.5.1869-1875.2000
Molecular differentiation of seven Malassezia species
Abstract
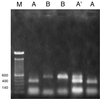
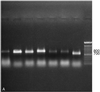
A system based on PCR and restriction endonuclease analysis was developed to distinguish the seven currently recognized Malassezia species. Seventy-eight strains, including authentic culture collection strains and routine clinical isolates, were investigated for variation in the ribosomal DNA repeat units. Two genomic regions, namely, the large subunit of the ribosomal gene and the internal transcribed spacer (ITS) region, were amplified by PCR, and products were digested with restriction endonucleases. The patterns generated were useful in identification of five out of seven Malassezia species. M. sympodialis was readily distinguishable in that its ITS region yielded a 700-bp amplified fragment, whereas the other six species yielded an 800-bp fragment. M. globosa and M. restricta were very similar in the regions studied and could be distinguished only by performing a hot start-touchdown PCR on primers for the beta-tubulin gene. Primers based on the conserved areas of the Candida cylindracea lipase gene, which were used in an attempt to amplify Malassezia lipases, yielded an amplification product after annealing at 55 degrees C only with M. pachydermatis. This specific amplification may facilitate the rapid identification of this organism.
Figures



References
-
- Anaissie E, Bodey G P. Nosocomial fungal infections: old problems and new challenges. Infect Dis Clin N Am. 1989;3:867–882. - PubMed
-
- Anthony R M, Howell S A, Pinters L. Application of DNA typing methods to the study of the epidemiology of Malassezia pachydermatis. Microb Ecol Health Dis. 1994;7:161–168.
-
- Ault G S, Ryschkewitsch C F, Stoner G L. Type-specific amplification of viral DNA using touchdown and hotstart PCR. J Virol Methods. 1994;46:145–156. - PubMed
-
- Boekhout T, Bosboom R W. Karyotyping of Malassezia yeasts: taxonomic and epidemiological implications. Syst Appl Microbiol. 1994;17:147–153.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

