Direct PCR of Cryptococcus neoformans MATalpha and MATa pheromones to determine mating type, ploidy, and variety: a tool for epidemiological and molecular pathogenesis studies
- PMID: 10790143
- PMCID: PMC86654
- DOI: 10.1128/JCM.38.5.2007-2009.2000
Direct PCR of Cryptococcus neoformans MATalpha and MATa pheromones to determine mating type, ploidy, and variety: a tool for epidemiological and molecular pathogenesis studies
Abstract
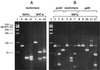
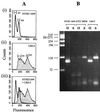
Cryptococcus neoformans MATalpha and MATa pheromones were amplified by direct PCR. Nucleotide sequence analyses revealed unique restriction enzyme sites. Sixty strains were used to devise a restriction fragment length polymorphism typing scheme that yielded three variety-specific patterns. Additionally, pheromone-specific PCR allowed easier identification of diploid C. neoformans strains than flow cytometry.
Figures
References
-
- Casadevall A, Perfect J R. Cryptococcus neoformans. Washington, D.C.: American Society for Microbiology; 1998.
-
- Heitman J, Allen B, Alspaugh J A, Kwon-Chung K J. On the origins of congenic MATα and MATa strains of the pathogenic yeast Cryptococcus neoformans. Fungal Genet Biol. 1999;28:1–5. - PubMed
-
- Huxley C, Green E D, Dunham I. Rapid assessment of Saccharomyces cerevisiae mating type by PCR. Trends Genet. 1990;6:236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

