GATA-dependent recruitment of MEF2 proteins to target promoters
- PMID: 10790371
- PMCID: PMC305697
- DOI: 10.1093/emboj/19.9.2046
GATA-dependent recruitment of MEF2 proteins to target promoters
Abstract
The myocyte enhancer factor-2 (MEF2) proteins are MADS-box transcription factors that are essential for differentiation of all muscle lineages but their mechanisms of action remain largely undefined. In mammals, the earliest site of MEF2 expression is the heart where the MEF2C isoform is detectable as early as embryonic day 7.5. Inactivation of the MEF2C gene causes cardiac developmental arrest and severe downregulation of a number of cardiac markers including atrial natriuretic factor (ANF). However, most of these promoters contain no or low affinity MEF2 binding sites and they are not significantly activated by any MEF2 proteins in heterologous cells suggesting a dependence on a cardiac-enriched cofactor for MEF2 action. We provide evidence that MEF2 proteins are recruited to target promoters by the cell-specific GATA transcription factors, and that MEF2 potentiates the transcriptional activity of this family of tissue-restricted zinc finger proteins. Functional MEF2/GATA-4 synergy involves physical interaction between the MEF2 DNA-binding domain and the carboxy zinc finger of GATA-4 and requires the activation domains of both proteins. However, neither MEF2 binding sites nor MEF2 DNA binding capacity are required for transcriptional synergy. The results unravel a novel pathway for transcriptional regulation by MEF2 and provide a molecular paradigm for elucidating the mechanisms of action of MEF2 in muscle and non-muscle cells.
Figures
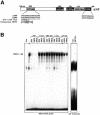
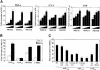
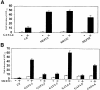
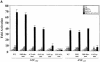




References
-
- Abdellatif M., MacLellan,W.R. and Schneider,M.D. (1994) p21 Ras as a governor of global gene expression. J. Biol. Chem., 269, 15423–15426. - PubMed
-
- Bi W., Drake,C.J. and Schwarz,J.J. (1999) The transcription factor MEF2C-null mouse exhibits complex vascular malformations and reduced cardiac expression of angiopoietin 1 and VEGF. Dev. Biol., 211, 255–267. - PubMed
-
- Black B.L. and Olson,E.N. (1998) Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell. Dev. Biol., 14, 167–196. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

