HRD gene dependence of endoplasmic reticulum-associated degradation
- PMID: 10793145
- PMCID: PMC14877
- DOI: 10.1091/mbc.11.5.1697
HRD gene dependence of endoplasmic reticulum-associated degradation
Abstract
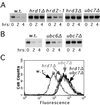
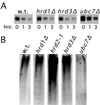
Work from several laboratories has indicated that many different proteins are subject to endoplasmic reticulum (ER) degradation by a common ER-associated machinery. This machinery includes ER membrane proteins Hrd1p/Der3p and Hrd3p and the ER-associated ubiquitin-conjugating enzymes Ubc7p and Ubc6p. The wide variety of substrates for this degradation pathway has led to the reasonable hypothesis that the HRD (Hmg CoA reductase degradation) gene-encoded proteins are generally involved in ER protein degradation in eukaryotes. We have tested this model by directly comparing the HRD dependency of the ER-associated degradation for various ER membrane proteins. Our data indicated that the role of HRD genes in protein degradation, even in this highly defined subset of proteins, can vary from absolute dependence to complete independence. Thus, ER-associated degradation can occur by mechanisms that do not involve Hrd1p or Hrd3p, despite their apparently broad envelope of substrates. These data favor models in which the HRD gene-encoded proteins function as specificity factors, such as ubiquitin ligases, rather than as factors involved in common aspects of ER degradation.
Figures

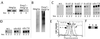
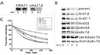
Similar articles
-
Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation.Nat Cell Biol. 2001 Jan;3(1):24-9. doi: 10.1038/35050524. Nat Cell Biol. 2001. PMID: 11146622
-
An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitination and ER-Golgi transport.J Cell Biol. 2002 Jul 8;158(1):91-101. doi: 10.1083/jcb.200201053. Epub 2002 Jul 8. J Cell Biol. 2002. PMID: 12105183 Free PMC article.
-
Sec61p-independent degradation of the tail-anchored ER membrane protein Ubc6p.EMBO J. 2001 Jun 15;20(12):3124-31. doi: 10.1093/emboj/20.12.3124. EMBO J. 2001. PMID: 11406589 Free PMC article.
-
Endoplasmic reticulum degradation: reverse protein flow of no return.FASEB J. 1997 Dec;11(14):1227-33. doi: 10.1096/fasebj.11.14.9409541. FASEB J. 1997. PMID: 9409541 Review.
-
The role of the ubiquitination machinery in dislocation and degradation of endoplasmic reticulum proteins.Curr Top Microbiol Immunol. 2005;300:57-93. doi: 10.1007/3-540-28007-3_4. Curr Top Microbiol Immunol. 2005. PMID: 16573237 Review.
Cited by
-
Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p.J Cell Biol. 2000 Oct 2;151(1):69-82. doi: 10.1083/jcb.151.1.69. J Cell Biol. 2000. PMID: 11018054 Free PMC article.
-
For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection.EMBO J. 2003 May 15;22(10):2309-17. doi: 10.1093/emboj/cdg227. EMBO J. 2003. PMID: 12743025 Free PMC article. Review.
-
E2-25K mediates US11-triggered retro-translocation of MHC class I heavy chains in a permeabilized cell system.Proc Natl Acad Sci U S A. 2006 Aug 1;103(31):11589-94. doi: 10.1073/pnas.0605215103. Epub 2006 Jul 25. Proc Natl Acad Sci U S A. 2006. PMID: 16868077 Free PMC article.
-
Insulin-induced gene protein (INSIG)-dependent sterol regulation of Hmg2 endoplasmic reticulum-associated degradation (ERAD) in yeast.J Biol Chem. 2013 Mar 22;288(12):8519-8530. doi: 10.1074/jbc.M112.404517. Epub 2013 Jan 10. J Biol Chem. 2013. PMID: 23306196 Free PMC article.
-
A protein complex containing Epo1p anchors the cortical endoplasmic reticulum to the yeast bud tip.J Cell Biol. 2015 Jan 5;208(1):71-87. doi: 10.1083/jcb.201407126. Epub 2014 Dec 29. J Cell Biol. 2015. PMID: 25547157 Free PMC article.
References
-
- Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in-vivo degradation of the yeast Mat-alpha-2 repressor. Cell. 1993;74:357–369. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

