Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway
- PMID: 10805719
- PMCID: PMC85689
- DOI: 10.1128/MCB.20.11.3753-3763.2000
Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway
Abstract
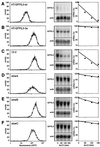
Control of mRNA stability is critical for expression of short-lived transcripts from cytokines and proto-oncogenes. Regulation involves an AU-rich element (ARE) in the 3' untranslated region (3'UTR) and cognate trans-acting factors thought to promote either degradation or stabilization of the mRNA. In this study we present a novel approach using somatic cell genetics designed to identify regulators of interleukin-3 (IL-3) mRNA turnover. Mutant cell lines were generated from diploid HT1080 cells transfected with a reporter construct containing green fluorescent protein (GFP) linked to the IL-3 3'UTR. GFP was expressed at low levels due to rapid decay of the mRNA. Following chemical mutagenesis and selection of GFP-overexpressing cells, we could isolate three mutant clones (slowA, slowB, and slowC) with a specific, trans-acting defect in IL-3 mRNA degradation, while the stability of IL-2 and tumor necrosis factor alpha reporter transcripts was not affected. Somatic cell fusion experiments revealed that the mutants are genetically recessive and form two complementation groups. Expression of the tristetraprolin gene in both groups led to reversion of the mutant phenotype, thereby linking this gene to the IL-3 mRNA degradation pathway. The genetic approach described here should allow identification of the defective functions by gene transfer and is also applicable to the study of other mRNA turnover pathways.
Figures





References
-
- Beelman C A, Parker R. Degradation of mRNA in eukaryotes. Cell. 1995;81:179–183. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
