Analysis of ku80-mutant mice and cells with deficient levels of p53
- PMID: 10805721
- PMCID: PMC85695
- DOI: 10.1128/MCB.20.11.3772-3780.2000
Analysis of ku80-mutant mice and cells with deficient levels of p53
Abstract
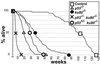
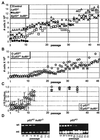
Absence of Ku80 results in increased sensitivity to ionizing radiation, defective lymphocyte development, early onset of an age-related phenotype, and premature replicative senescence. Here we investigate the role of p53 on the phenotype of ku80-mutant mice and cells. Reducing levels of p53 increased the cancer incidence for ku80(-/-) mice. About 20% of ku80(-/-) p53(+/-) mice developed a broad spectrum of cancer by 40 weeks and all ku80(-/-) p53(-/-) mice developed pro-B-cell lymphoma by 16 weeks. Reducing levels of p53 rescued populations of ku80(-/-) cells from replicative senescence by enabling spontaneous immortalization. The double-mutant cells are impaired for the G(1)/S checkpoint due to the p53 mutation and are hypersensitive to gamma-radiation and reactive oxygen species due to the Ku80 mutation. These data show that replicative senescence is caused by a p53-dependent cell cycle response to damaged DNA in ku80(-/-) cells and that p53 is essential for preventing very early onset of pro-B-cell lymphoma in ku80(-/-) mice.
Figures




Similar articles
-
Ku80 and p53 suppress medulloblastoma that arise independent of Rag-1-induced DSBs.Oncogene. 2006 Nov 16;25(54):7159-65. doi: 10.1038/sj.onc.1209704. Epub 2006 Jun 5. Oncogene. 2006. PMID: 16751807
-
Unlike p53, p27 failed to exhibit an anti-tumor genetic interaction with Ku80.Cell Cycle. 2009 Aug;8(15):2463-6. doi: 10.4161/cc.8.15.9249. Epub 2009 Aug 11. Cell Cycle. 2009. PMID: 19597334
-
Deleting Ku70 is milder than deleting Ku80 in p53-mutant mice and cells.Oncogene. 2009 Apr 23;28(16):1875-8. doi: 10.1038/onc.2009.57. Epub 2009 Mar 30. Oncogene. 2009. PMID: 19330025
-
DNA repair factors and telomere-chromosome integrity in mammalian cells.Cytogenet Genome Res. 2004;104(1-4):116-22. doi: 10.1159/000077475. Cytogenet Genome Res. 2004. PMID: 15162024 Review.
-
Ku, a DNA repair protein with multiple cellular functions?Mutat Res. 1999 May 14;434(1):3-15. doi: 10.1016/s0921-8777(99)00006-3. Mutat Res. 1999. PMID: 10377944 Review.
Cited by
-
HIV-1 Tat and AIDS-associated cancer: targeting the cellular anti-cancer barrier?J Exp Clin Cancer Res. 2008 May 15;27(1):3. doi: 10.1186/1756-9966-27-3. J Exp Clin Cancer Res. 2008. PMID: 18577246 Free PMC article. Review.
-
Effect of Ku80 deficiency on mutation frequencies and spectra at a LacZ reporter locus in mouse tissues and cells.PLoS One. 2008;3(10):e3458. doi: 10.1371/journal.pone.0003458. Epub 2008 Oct 20. PLoS One. 2008. PMID: 18941635 Free PMC article.
-
Senescence and aging: Causes, consequences, and therapeutic avenues.J Cell Biol. 2018 Jan 2;217(1):65-77. doi: 10.1083/jcb.201708092. Epub 2017 Nov 7. J Cell Biol. 2018. PMID: 29114066 Free PMC article. Review.
-
Effects of p21 deletion in mouse models of premature aging.Cell Cycle. 2009 Jul 1;8(13):2002-4. doi: 10.4161/cc.8.13.8997. Epub 2009 Jul 12. Cell Cycle. 2009. PMID: 19535900 Free PMC article.
-
DNA damage responses and p53 in the aging process.Blood. 2018 Feb 1;131(5):488-495. doi: 10.1182/blood-2017-07-746396. Epub 2017 Nov 15. Blood. 2018. PMID: 29141944 Free PMC article. Review.
References
-
- Bernstein C, Bernstein H. Aging, sex, and DNA repair. San Diego, Calif: Academic Press; 1991.
-
- Bogue M A, Wang C, Zhu C, Roth D B. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity. 1997;7:37–47. - PubMed
-
- Bogue M A, Zhu C, Aguilar-Cordova E, Donehower L A, Roth D B. p53 is required for both radiation-induced differentiation and rescue of V(D)J rearrangement in scid mouse thymocytes. Genes Dev. 1996;10:1–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
