Prolyl 4-hydroxylase is an essential procollagen-modifying enzyme required for exoskeleton formation and the maintenance of body shape in the nematode Caenorhabditis elegans
- PMID: 10805750
- PMCID: PMC85778
- DOI: 10.1128/MCB.20.11.4084-4093.2000
Prolyl 4-hydroxylase is an essential procollagen-modifying enzyme required for exoskeleton formation and the maintenance of body shape in the nematode Caenorhabditis elegans
Abstract
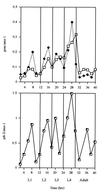
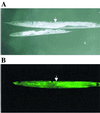
The multienzyme complex prolyl 4-hydroxylase catalyzes the hydroxylation of proline residues and acts as a chaperone during collagen synthesis in multicellular organisms. The beta subunit of this complex is identical to protein disulfide isomerase (PDI). The free-living nematode Caenorhabditis elegans is encased in a collagenous exoskeleton and represents an excellent model for the study of collagen biosynthesis and extracellular matrix formation. In this study, we examined prolyl 4-hydroxylase alpha-subunit (PHY; EC 1.14.11.2)- and beta-subunit (PDI; EC 5.3.4.1)-encoding genes with respect to their role in collagen modification and formation of the C. elegans exoskeleton. We identified genes encoding two PHYs and a single associated PDI and showed that all three are expressed in collagen-synthesizing ectodermal cells at times of maximal collagen synthesis. Disruption of the pdi gene via RNA interference resulted in embryonic lethality. Similarly, the combined phy genes are required for embryonic development. Interference with phy-1 resulted in a morphologically dumpy phenotype, which we determined to be identical to the uncharacterized dpy-18 locus. Two dpy-18 mutant strains were shown to have null alleles for phy-1 and to have a reduced hydroxyproline content in their exoskeleton collagens. This study demonstrates in vivo that this enzyme complex plays a central role in extracellular matrix formation and is essential for normal metazoan development.
Figures




References
-
- Annunen P, Helaakoski T, Myllyharju J, Veijola J, Pihlajaniemi T, Kivirikko K I. Cloning of the human prolyl 4-hydroxylase alpha subunit isoform alpha(II) and characterization of the type II enzyme tetramer—the alpha(I) and alpha(II) subunits do not form a mixed alpha(I)alpha(II)beta(2) tetramer. J Biol Chem. 1997;272:17342–17348. - PubMed
-
- Annunen P, Koivunen P, Kivirikko K I. Cloning of the alpha subunit of prolyl 4-hydroxylase from Drosophila and expression and characterization of the corresponding enzyme tetramer with some unique properties. J Biol Chem. 1999;274:6790–6796. - PubMed
-
- Blelloch R, Kimble J. Control of organ shape by a secreted metalloprotease in the nematode Caenorhabditis elegans. Nature. 1999;399:586–590. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
