An ancestral MADS-box gene duplication occurred before the divergence of plants and animals
- PMID: 10805792
- PMCID: PMC25828
- DOI: 10.1073/pnas.97.10.5328
An ancestral MADS-box gene duplication occurred before the divergence of plants and animals
Abstract
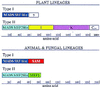
Changes in genes encoding transcriptional regulators can alter development and are important components of the molecular mechanisms of morphological evolution. MADS-box genes encode transcriptional regulators of diverse and important biological functions. In plants, MADS-box genes regulate flower, fruit, leaf, and root development. Recent sequencing efforts in Arabidopsis have allowed a nearly complete sampling of the MADS-box gene family from a single plant, something that was lacking in previous phylogenetic studies. To test the long-suspected parallel between the evolution of the MADS-box gene family and the evolution of plant form, a polarized gene phylogeny is necessary. Here we suggest that a gene duplication ancestral to the divergence of plants and animals gave rise to two main lineages of MADS-box genes: TypeI and TypeII. We locate the root of the eukaryotic MADS-box gene family between these two lineages. A novel monophyletic group of plant MADS domains (AGL34 like) seems to be more closely related to previously identified animal SRF-like MADS domains to form TypeI lineage. Most other plant sequences form a clear monophyletic group with animal MEF2-like domains to form TypeII lineage. Only plant TypeII members have a K domain that is downstream of the MADS domain in most plant members previously identified. This suggests that the K domain evolved after the duplication that gave rise to the two lineages. Finally, a group of intermediate plant sequences could be the result of recombination events. These analyses may guide the search for MADS-box sequences in basal eukaryotes and the phylogenetic placement of new genes from other plant species.
Figures



References
-
- Shore P, Sharrocks A D. Eur J Biochem. 1995;229:1–13. - PubMed
-
- Coen E S, Meyerowitz E M. Nature (London) 1991;353:31–37. - PubMed
-
- Riechmann J L, Meyerowitz E M. J Biol Chem. 1997;378:1079–1101. - PubMed
-
- Liljegren S J, Ferrándiz C, Alvarez-Buylla E R, Pelaz S, Yanofsky M F. Flowering Newsletter. 1998;25:9–19.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

