Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections
- PMID: 10805806
- PMCID: PMC25859
- DOI: 10.1073/pnas.080520697
Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections
Abstract
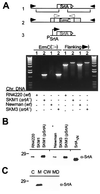
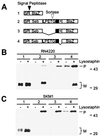
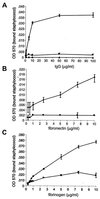
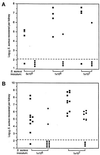
Many gram-positive bacteria covalently tether their surface adhesins to the cell wall peptidoglycan. We find that surface proteins of Staphylococcus aureus are linked to the cell wall by sortase, an enzyme that cleaves polypeptides at a conserved LPXTG motif. S. aureus mutants lacking sortase fail to process and display surface proteins and are defective in the establishment of infections. Thus, the cell wall envelope of gram-positive bacteria represents a surface organelle responsible for interactions with the host environment during the pathogenesis of bacterial infections.
Figures


Comment in
-
Sortase, a universal target for therapeutic agents against gram-positive bacteria?Proc Natl Acad Sci U S A. 2000 May 9;97(10):5013-5. doi: 10.1073/pnas.97.10.5013. Proc Natl Acad Sci U S A. 2000. PMID: 10805759 Free PMC article. Review. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

