Comparative analysis of expressed sequences in Phytophthora sojae
- PMID: 10806241
- PMCID: PMC58998
- DOI: 10.1104/pp.123.1.243
Comparative analysis of expressed sequences in Phytophthora sojae
Abstract
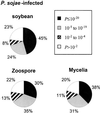
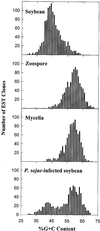
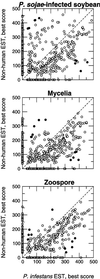
Phytophthora sojae (Kaufmann and Gerdemann) is an oomycete that causes stem and root rot on soybean (Glycine max L. Merr) plants. We have constructed three cDNA libraries using mRNA isolated from axenically grown mycelium and zoospores and from tissue isolated from plant hypocotyls 48 h after inoculation with zoospores. A total of 3,035 expressed sequence tags (ESTs) were generated from the three cDNA libraries, representing an estimated 2,189 cDNA transcripts. The ESTs were classified according to putative function based on similarity to known proteins, and were analyzed for redundancy within and among the three source libraries. Distinct expression patterns were observed for each library. By analysis of the percentage G+C content of the ESTs, we estimate that two-thirds of the ESTs from the infected plant library are derived from P. sojae cDNA transcripts. The ESTs originating from this study were also compared with a collection of Phytophthora infestans ESTs and with all other non-human ESTs to assess the similarity of the P. sojae sequences to existing EST data. This collection of cDNA libraries, ESTs, and accompanying annotation will provide a new resource for studies on oomycetes and on soybean responses to pathogen challenge.
Figures


References
-
- Adams MD, Kelley JM, Gocayne JD, Dubnick M, Polymeropoulos MH, Xiao H, Merril CR, Wu A, Olde B, Moreno RF, Kerlavage AR, McCombie WR, Venter JC. Complementary DNA sequencing: expressed sequence tags and human genome project. Science. 1991;252:1651–1656. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Anderson TR, Buzzell RI. Diversity and frequency of races of Phytophthora megasperma f. sp. glycinea in soybean fields in Essex County, Ontario, 1980–1989. Plant Dis. 1992;76:587–589.
-
- Dellaporta S, Woods H, Hicks J. A plant DNA mini-preparation: version II. Plant Mol Biol Rep. 1983;1:19–21.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

