Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions
- PMID: 10809668
- PMCID: PMC316578
Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions
Abstract
We provide direct evidence that pre-mRNA splicing alters mRNP protein composition. Using a novel in vitro cross-linking approach, we detected several proteins that associate with mRNA exon-exon junctions only as a consequence of splicing. Immunoprecipitation experiments suggested that these proteins are part of a tight complex around the junction. Two were identified as SRm160, a nuclear matrix-associated splicing coactivator, and hPrp8p, a core component of U5 snRNP and spliceosomes. Glycerol gradient fractionation showed that a subset of these proteins remain associated with mRNA after its release from the spliceosome. These results demonstrate that the spliceosome can leave behind signature proteins at exon-exon junctions. Such proteins could influence downstream metabolic events in vivo such as mRNA transport, translation, and nonsense-mediated decay.
Figures



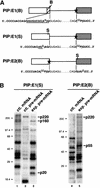

References
-
- Abmayr SM, Workman JL, Roeder RG. The pseudorabies immediate early protein stimulates in vitro transcription by facilitating TFIID: Promoter interactions. Genes & Dev. 1988;2:542–553. - PubMed
-
- Achsel T, Ahrens K, Brahms H, Teigelkamp S, Luhrmann R. The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol Cell Biol. 1998;18:6756–6766. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
