Expression and characterization of the chitin-binding domain of chitinase A1 from Bacillus circulans WL-12
- PMID: 10809681
- PMCID: PMC94488
- DOI: 10.1128/JB.182.11.3045-3054.2000
Expression and characterization of the chitin-binding domain of chitinase A1 from Bacillus circulans WL-12
Abstract
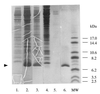
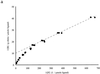
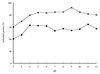
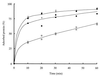
Chitinase A1 from Bacillus circulans WL-12 comprises an N-terminal catalytic domain, two fibronectin type III-like domains, and a C-terminal chitin-binding domain (ChBD). In order to study the biochemical properties and structure of the ChBD, ChBD(ChiA1) was produced in Escherichia coli using a pET expression system and purified by chitin affinity column chromatography. Purified ChBD(ChiA1) specifically bound to various forms of insoluble chitin but not to other polysaccharides, including chitosan, cellulose, and starch. Interaction of soluble chitinous substrates with ChBD(ChiA1) was not detected by means of nuclear magnetic resonance and isothermal titration calorimetry. In addition, the presence of soluble substrates did not interfere with the binding of ChBD(ChiA1) to regenerated chitin. These observations suggest that ChBD(ChiA1) recognizes a structure which is present in insoluble or crystalline chitin but not in chito-oligosaccharides or in soluble derivatives of chitin. ChBD(ChiA1) exhibited binding activity over a wide range of pHs, and the binding activity was enhanced at pHs near its pI and by the presence of NaCl, suggesting that the binding of ChBD(ChiA1) is mediated mainly by hydrophobic interactions. Hydrolysis of beta-chitin microcrystals by intact chitinase A1 and by a deletion derivative lacking the ChBD suggested that the ChBD is not absolutely required for hydrolysis of beta-chitin microcrystals but greatly enhances the efficiency of degradation.
Figures




Similar articles
-
Solution structure of the chitin-binding domain of Bacillus circulans WL-12 chitinase A1.J Biol Chem. 2000 May 5;275(18):13654-61. doi: 10.1074/jbc.275.18.13654. J Biol Chem. 2000. PMID: 10788483
-
Involvement of Gln679, in addition to Trp687, in chitin-binding activity of the chitin-binding domain of chitinase A1 from Bacillus circulans WL-12.J Biochem. 2013 Aug;154(2):185-93. doi: 10.1093/jb/mvt043. Epub 2013 May 20. J Biochem. 2013. PMID: 23694779
-
Identification of the substrate interaction region of the chitin-binding domain of Streptomyces griseus chitinase C.J Biochem. 2006 Mar;139(3):483-93. doi: 10.1093/jb/mvj062. J Biochem. 2006. PMID: 16567413
-
Chitins and chitosans as immunoadjuvants and non-allergenic drug carriers.Mar Drugs. 2010 Feb 21;8(2):292-312. doi: 10.3390/md8020292. Mar Drugs. 2010. PMID: 20390107 Free PMC article. Review.
-
Chitinase-Assisted Bioconversion of Chitinous Waste for Development of Value-Added Chito-Oligosaccharides Products.Biology (Basel). 2023 Jan 5;12(1):87. doi: 10.3390/biology12010087. Biology (Basel). 2023. PMID: 36671779 Free PMC article. Review.
Cited by
-
Fabrication of silica on chitin in ambient conditions using silicatein fused with a chitin-binding domain.Bioprocess Biosyst Eng. 2021 Sep;44(9):1883-1890. doi: 10.1007/s00449-021-02568-w. Epub 2021 May 11. Bioprocess Biosyst Eng. 2021. PMID: 33974134
-
Biodegradable plastics from renewable sources.Folia Microbiol (Praha). 2003;48(1):27-44. doi: 10.1007/BF02931273. Folia Microbiol (Praha). 2003. PMID: 12744074 Review.
-
Chitinase genes LbCHI31 and LbCHI32 from Limonium bicolor were successfully expressed in Escherichia coli and exhibit recombinant chitinase activities.ScientificWorldJournal. 2013 Dec 7;2013:648382. doi: 10.1155/2013/648382. eCollection 2013. ScientificWorldJournal. 2013. PMID: 24385885 Free PMC article.
-
Accelerated CO₂ Hydration with Thermostable Sulfurihydrogenibium azorense Carbonic Anhydrase-Chitin Binding Domain Fusion Protein Immobilised on Chitin Support.Int J Mol Sci. 2019 Mar 25;20(6):1494. doi: 10.3390/ijms20061494. Int J Mol Sci. 2019. PMID: 30934614 Free PMC article.
-
Potential role of chitinases and chitin-binding proteins in host-microbial interactions during the development of intestinal inflammation.Histol Histopathol. 2011 Nov;26(11):1453-64. doi: 10.14670/HH-26.1453. Histol Histopathol. 2011. PMID: 21938682 Free PMC article. Review.
References
-
- Alam M M, Nikaidou N, Tanaka H, Watanabe T. Cloning and sequencing of chiC gene of Bacillus circulans WL-12 and relationship of its product to some other chitinases and chitinase-like proteins. J Ferment Bioeng. 1995;80:454–461.
-
- Alam M M, Mizutani T, Isono M, Nikaidou N, Watanabe T. Three chitinase genes (chiA, chiC, and chiD) comprise the chitinase system of Bacillus circulans WL-12. J Ferment Bioeng. 1996;82:28–36.
-
- Brun E, Moriaud F, Gans P, Blackledge M J, Barras F, Marion D. Solution structure of the cellulose-binding domain of the endoglucanase Z secreted by Erwinia chrysanthemi. Biochemistry. 1997;36:16074–16086. - PubMed
-
- Chong S, Mersha F B, Comb D G, Scott M E, Landry D, Vence L M, Perler F B, Benner J, Kucera R B, Hirvonen C A, Pelletier J J, Paulus H, Xu M Q. Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene. 1997;192:271–281. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

