Fermentative metabolism of Bacillus subtilis: physiology and regulation of gene expression
- PMID: 10809684
- PMCID: PMC94491
- DOI: 10.1128/JB.182.11.3072-3080.2000
Fermentative metabolism of Bacillus subtilis: physiology and regulation of gene expression
Abstract
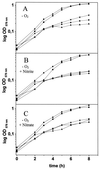
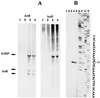
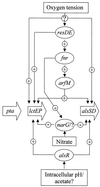
Bacillus subtilis grows in the absence of oxygen using nitrate ammonification and various fermentation processes. Lactate, acetate, and 2,3-butanediol were identified in the growth medium as the major anaerobic fermentation products by using high-performance liquid chromatography. Lactate formation was found to be dependent on the lctEP locus, encoding lactate dehydrogenase and a putative lactate permease. Mutation of lctE results in drastically reduced anaerobic growth independent of the presence of alternative electron acceptors, indicating the importance of NADH reoxidation by lactate dehydrogenase for the overall anaerobic energy metabolism. Anaerobic formation of 2,3-butanediol via acetoin involves acetolactate synthase and decarboxylase encoded by the alsSD operon. Mutation of alsSD has no significant effect on anaerobic growth. Anaerobic acetate synthesis from acetyl coenzyme A requires phosphotransacetylase encoded by pta. Similar to the case for lctEP, mutation of pta significantly reduces anaerobic fermentative and respiratory growth. The expression of both lctEP and alsSD is strongly induced under anaerobic conditions. Anaerobic lctEP and alsSD induction was found to be partially dependent on the gene encoding the redox regulator Fnr. The observed fnr dependence might be the result of Fnr-induced arfM (ywiD) transcription and subsequent lctEP and alsSD activation by the regulator ArfM (YwiD). The two-component regulatory system encoded by resDE is also involved in anaerobic lctEP induction. No direct resDE influence on the redox regulation of alsSD was observed. The alternative electron acceptor nitrate represses anaerobic lctEP and alsSD transcription. Nitrate repression requires resDE- and fnr-dependent expression of narGHJI, encoding respiratory nitrate reductase. The gene alsR, encoding a regulator potentially responding to changes of the intracellular pH and to acetate, is essential for anaerobic lctEP and alsSD expression. In agreement with its known aerobic function, no obvious oxygen- or nitrate-dependent pta regulation was observed. A model for the regulation of the anaerobic fermentation genes in B. subtilis is proposed.
Figures


Similar articles
-
Modulation of anaerobic energy metabolism of Bacillus subtilis by arfM (ywiD).J Bacteriol. 2001 Dec;183(23):6815-21. doi: 10.1128/JB.183.23.6815-6821.2001. J Bacteriol. 2001. PMID: 11698370 Free PMC article.
-
The transcription factor AlsR binds and regulates the promoter of the alsSD operon responsible for acetoin formation in Bacillus subtilis.J Bacteriol. 2012 Mar;194(5):1100-12. doi: 10.1128/JB.06425-11. Epub 2011 Dec 16. J Bacteriol. 2012. PMID: 22178965 Free PMC article.
-
Characterization of anaerobic fermentative growth of Bacillus subtilis: identification of fermentation end products and genes required for growth.J Bacteriol. 1997 Nov;179(21):6749-55. doi: 10.1128/jb.179.21.6749-6755.1997. J Bacteriol. 1997. PMID: 9352926 Free PMC article.
-
Regulation of the anaerobic metabolism in Bacillus subtilis.Adv Microb Physiol. 2012;61:195-216. doi: 10.1016/B978-0-12-394423-8.00005-6. Adv Microb Physiol. 2012. PMID: 23046954 Review.
-
Anaerobic growth of a "strict aerobe" (Bacillus subtilis).Annu Rev Microbiol. 1998;52:165-90. doi: 10.1146/annurev.micro.52.1.165. Annu Rev Microbiol. 1998. PMID: 9891797 Review.
Cited by
-
Integrative Analysis of Proteome and Transcriptome Dynamics during Bacillus subtilis Spore Revival.mSphere. 2020 Aug 5;5(4):e00463-20. doi: 10.1128/mSphere.00463-20. mSphere. 2020. PMID: 32759332 Free PMC article.
-
Acetic Acid Acts as a Volatile Signal To Stimulate Bacterial Biofilm Formation.mBio. 2015 Jun 9;6(3):e00392. doi: 10.1128/mBio.00392-15. mBio. 2015. PMID: 26060272 Free PMC article.
-
Anode-assisted electro-fermentation with Bacillus subtilis under oxygen-limited conditions.Biotechnol Biofuels Bioprod. 2023 Jan 10;16(1):6. doi: 10.1186/s13068-022-02253-4. Biotechnol Biofuels Bioprod. 2023. PMID: 36627716 Free PMC article.
-
Roles of d-Lactate Dehydrogenases in the Anaerobic Growth of Shewanella oneidensis MR-1 on Sugars.Appl Environ Microbiol. 2019 Jan 23;85(3):e02668-18. doi: 10.1128/AEM.02668-18. Print 2019 Feb 1. Appl Environ Microbiol. 2019. PMID: 30504209 Free PMC article.
-
Distinct effects of sorbic acid and acetic acid on the electrophysiology and metabolism of Bacillus subtilis.Appl Environ Microbiol. 2014 Oct;80(19):5918-26. doi: 10.1128/AEM.01391-14. Epub 2014 Jul 18. Appl Environ Microbiol. 2014. PMID: 25038097 Free PMC article.
References
-
- Antelmann H, Bernhardt J, Schmid R, Mach H, Volker U, Hecker M. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis. 1997;18:1451–1463. - PubMed
-
- Böck A, Sawers G. Fermentation. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 262–282.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

