Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus
- PMID: 10809700
- PMCID: PMC94507
- DOI: 10.1128/JB.182.11.3197-3203.2000
Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus
Abstract
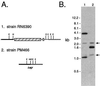
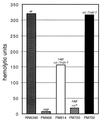
A chromosomal insertion of transposon Tn917 partially restores the expression of protease and alpha-toxin activities to PM466, a genetically defined agr-null derivative of the wild-type Staphylococcus aureus strain RN6390. In co-transduction experiments, transposon-encoded erythromycin resistance and a protease- and alpha-toxin-positive phenotype are transferred at high frequency from mutant strains to agr-null strains of S. aureus. Southern analysis of chromosomal DNA and sequence analysis of DNA flanking the Tn917 insertion site in mutant strains revealed that the transposon interrupted a 498-bp open reading frame (ORF). Similarity searches using a conceptual translation of the ORF identified a region of homology to the known staphylococcal global regulators AgrA and SarA. To verify that the mutant allele conferred the observed phenotype, a wild-type allele of the mutant gene was introduced into the genome of a mutant strain by homologous recombination. The resulting isolates had a restored agr-null phenotype. Virulence factor gene expression in mutant, restored mutant, and wild-type strains was quantified by measuring alpha-toxin activity in culture supernatant fluids and by Northern analysis of the alpha-toxin transcript. We named this ORF rot (for repressor of toxins) (GenBank accession no. AF189239) because of the activity associated with rot::Tn917 mutant strains.
Figures
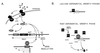


Similar articles
-
SarT, a repressor of alpha-hemolysin in Staphylococcus aureus.Infect Immun. 2001 Aug;69(8):4749-58. doi: 10.1128/IAI.69.8.4749-4758.2001. Infect Immun. 2001. PMID: 11447147 Free PMC article.
-
Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus.J Bacteriol. 1998 Dec;180(23):6232-41. doi: 10.1128/JB.180.23.6232-6241.1998. J Bacteriol. 1998. PMID: 9829932 Free PMC article.
-
Global regulation of Staphylococcus aureus genes by Rot.J Bacteriol. 2003 Jan;185(2):610-9. doi: 10.1128/JB.185.2.610-619.2003. J Bacteriol. 2003. PMID: 12511508 Free PMC article.
-
Identification of single nucleotide polymorphisms associated with hyperproduction of alpha-toxin in Staphylococcus aureus.PLoS One. 2011 Apr 8;6(4):e18428. doi: 10.1371/journal.pone.0018428. PLoS One. 2011. PMID: 21494631 Free PMC article.
-
Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family.Front Biosci. 2002 Aug 1;7:d1825-42. doi: 10.2741/A882. Front Biosci. 2002. PMID: 12133812 Review.
Cited by
-
sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4.J Bacteriol. 2002 Oct;184(19):5457-67. doi: 10.1128/JB.184.19.5457-5467.2002. J Bacteriol. 2002. PMID: 12218034 Free PMC article.
-
Staphylococcus aureus isolated in cases of impetigo produces both epidermolysin A or B and LukE-LukD in 78% of 131 retrospective and prospective cases.J Clin Microbiol. 2001 Dec;39(12):4349-56. doi: 10.1128/JCM.39.12.4349-4356.2001. J Clin Microbiol. 2001. PMID: 11724844 Free PMC article.
-
Mgr, a novel global regulator in Staphylococcus aureus.J Bacteriol. 2003 Jul;185(13):3703-10. doi: 10.1128/JB.185.13.3703-3710.2003. J Bacteriol. 2003. PMID: 12813062 Free PMC article.
-
SarS Is a Repressor of Staphylococcus aureus Bicomponent Pore-Forming Leukocidins.Infect Immun. 2023 Apr 18;91(4):e0053222. doi: 10.1128/iai.00532-22. Epub 2023 Mar 20. Infect Immun. 2023. PMID: 36939325 Free PMC article.
-
sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus.Infect Immun. 2003 Jan;71(1):343-53. doi: 10.1128/IAI.71.1.343-353.2003. Infect Immun. 2003. PMID: 12496184 Free PMC article.
References
-
- Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular Biology. New York, N.Y: Greene Publishing and Wiley-Interscience; 1996.
-
- Balaban N, Novick R P. Translation of RNAIII, the Staphylococcus aureus agr regulatory RNA molecule, can be activated by a 3′-end deletion. FEMS Microbiol Lett. 1995;133:155–161. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

