MHC class II pseudogene and genomic signature of a 32-kb cosmid in the house finch (Carpodacus mexicanus)
- PMID: 10810083
- PMCID: PMC310861
- DOI: 10.1101/gr.10.5.613
MHC class II pseudogene and genomic signature of a 32-kb cosmid in the house finch (Carpodacus mexicanus)
Abstract
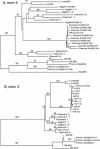
Large-scale sequencing studies in vertebrates have thus far focused primarily on the genomes of a few model organisms. Birds are of interest to genomics because of their much smaller and highly streamlined genomes compared to mammals. However, large-scale genetic work has been confined almost exclusively to the chicken; we know little about general aspects of genomes in nongame birds. This study examines the organization of a genomic region containing an Mhc class II B gene in a representative of another important lineage of the avian tree, the songbirds (Passeriformes). We used a shotgun sequencing approach to determine the sequence of a 32-kb cosmid insert containing a strongly hybridizing Mhc fragment from house finches (Carpodacus mexicanus). There were a total of three genes found on the cosmid clone, about the gene density expected for the mammalian Mhc: a class II Mhc beta-chain gene (Came-DAB1), a serine-threonine kinase, and a zinc finger motif. Frameshift mutations in both the second and third exons of Came-DAB1 and the unalignability of the gene after the third exon suggest that it is a nonfunctional pseudogene. In addition, the identifiable introns of Came-DAB1 are more than twice as large as those of chickens. Nucleotide diversity in the peptide-binding region of Came-DAB1 (Pi = 0.03) was much lower than polymorphic chicken and other functional Mhc genes but higher than the expected diversity for a neutral locus in birds, perhaps because of hitchhiking on a selected Mhc locus close by. The serine-threonine kinase gene is likely functional, whereas the zinc finger motif is likely nonfunctional. A paucity of long simple-sequence repeats and retroelements is consistent with emerging rules of chicken genomics, and a pictorial analysis of the "genomic signature" of this sequence, the first of its kind for birds, bears strong similarity to mammalian signatures, suggesting common higher-order structures in these homeothermic genomes. The house finch sequence is among a very few of its kind from nonmodel vertebrates and provides insight into the evolution of the avian Mhc and of avian genomes generally.
Figures

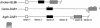


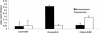

References
-
- Becker KG, Nagle JW, Cannin RD, Biddison WE, Ozato K, Drew PD. Rapid isolation and characterization of 118 novel C2H2-type zinc finger cDNAs expressed in human brain. Hum Mol Genet. 1995;4:685–691. - PubMed
-
- Beckmann JS, Weber JL. Survey of human and rat microsatellites. Genomics. 1992;12:627–631. - PubMed
-
- Bernardi G, Hughes S, Mouchiroud D. The major compositional transitions in the vertebrate genome. J Mol Evol. 1997;44:S44–S51. - PubMed
-
- Bingulac-Popovic J, Figueroa F, Sato A, Talbot WS, Johnson SL, Gates M, Postlethwait JH, Klein J. Mapping of Mhc class I and class II regions to different linkage groups in the zebrafish, Danio rerio. Immunogenetics. 1997;46:129–134. - PubMed
-
- Briles WE, McGibbon WH. Heterozygosity of inbred lines of chickens at two loci affecting cellular antigens. Genetics. 1948;33:605. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
