A radiation hybrid map of the cat genome: implications for comparative mapping
- PMID: 10810092
- PMCID: PMC310870
- DOI: 10.1101/gr.10.5.691
A radiation hybrid map of the cat genome: implications for comparative mapping
Abstract
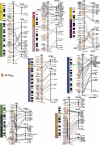
Ordered gene maps of mammalian species are becoming increasingly valued in assigning gene variants to function in human and animal models, as well as recapitulating the natural history of genome organization. To extend this power to the domestic cat, a radiation hybrid (RH) map of the cat was constructed integrating 424 Type I-coding genes with 176 microsatellite markers, providing coverage over all 20 feline chromosomes. Alignment of parallel RH maps of human and cat reveal 100 conserved segments ordered (CSOs) between the species, nearly three times the number observed with reciprocal chromosome painting analyses. The observed number is equivalent to theoretical predictions of the number of conserved segments to be found between cat and human, implying that 300-400 Type I gene markers is sufficient to reveal nearly all conserved segments for species that exhibit the most frequently observed "slow" rate of genome reorganization. The cat-human RH map comparisons provide a new genomic tool for comparative gene mapping in the cat and related Felidae, and provide confirmation that the cat genome organization is remarkably conserved compared with human. These data demonstrate that ordered RH-based gene maps provide the most precise assessment of comparing genomes, short of contig construction or full-sequence determination.
Figures



References
-
- Andersson L, Archibald A, Ashburner M, Audun S, Barendse W, Bitgood J, Bottema C, Broad T, Brown S, Burt D, et al. Comparative genome organization of vertebrates. The first international workshop on comparative genome organization. Mamm Genome. 1996;7:717–734. - PubMed
-
- Bielec PE, Gallagher DS, Womack JE, Busbee DL. Homologies between human and dolphin chromosomes detected by heterologous chromosome painting. Cytogenet Cell Genet. 1998;81:18–25. - PubMed
-
- Biller DS, DiBartola SP, Eaton KA, Pflueger S, Wellman ML, Radin MJ. Inheritance of polycystic kidney disease in Persian cats. J Hered. 1996;87:1–5. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
