Genome relationships: the grass model in current research
- PMID: 10810140
- PMCID: PMC139917
- DOI: 10.1105/tpc.12.5.637
Genome relationships: the grass model in current research
Figures


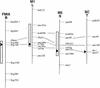
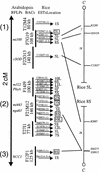
References
-
- Büschges, R., et al. (1997). The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 88, 695–705. - PubMed
-
- Chao, S., Sharp, P.J., Worland, A.J., Warham, E.J., Koebner, R.M.D., and Gale, M.D. (1989). RFLP-based genetic maps of wheat homoeologous group 7 chromosomes. Theor. Appl. Genet. 78, 495–504. - PubMed
-
- Devos, K.M., Atkinson, M.D., Chinoy, C.N., Harcourt, R.L., Koebner, R.M.D., Liu, C.J., Masojc, P., Xie, D.X., and Gale, M.D. (1993. a). Chromosomal rearrangements in the rye genome relative to that of wheat. Theor. Appl. Genet. 85, 673–680. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

