Virus-induced silencing of a plant cellulose synthase gene
- PMID: 10810144
- PMCID: PMC139921
- DOI: 10.1105/tpc.12.5.691
Virus-induced silencing of a plant cellulose synthase gene
Abstract
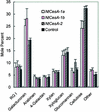
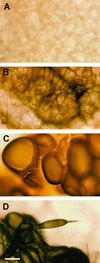
Specific cDNA fragments corresponding to putative cellulose synthase genes (CesA) were inserted into potato virus X vectors for functional analysis in Nicotiana benthamiana by using virus-induced gene silencing. Plants infected with one group of cDNAs had much shorter internode lengths, small leaves, and a "dwarf" phenotype. Consistent with a loss of cell wall cellulose, abnormally large and in many cases spherical cells ballooned from the undersurfaces of leaves, particularly in regions adjacent to vascular tissues. Linkage analyses of wall polysaccharides prepared from infected leaves revealed a 25% decrease in cellulose content. Transcript levels for at least one member of the CesA cellulose synthase gene family were lower in infected plants. The decrease in cellulose content in cell walls was offset by an increase in homogalacturonan, in which the degree of esterification of carboxyl groups decreased from approximately 50 to approximately 33%. The results suggest that feedback loops interconnect the cellular machinery controlling cellulose and pectin biosynthesis. On the basis of the phenotypic features of the infected plants, changes in wall composition, and the reduced abundance of CesA mRNA, we concluded that the cDNA fragments silenced one or more cellulose synthase genes.
Figures




References
-
- Arioli, T., et al. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279, 717–720. - PubMed
-
- Bacic, A., Harris, P.J., and Stone, B.A. (1988). Structure and function of plant cell walls. In The Biochemistry of Plants: A Comprehensive Treatise, Vol. 14, Carbohydrates, J. Preiss, ed (New York: Academic Press), pp. 297–371.
-
- Baulcombe, D.C. (1999). Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 2, 109–113. - PubMed
-
- Baulcombe, D.C., Chapman, S., and Cruz, S.S. (1995). Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 7, 1045–1053. - PubMed
-
- Brett, C., and Waldron, K. (1990). Physiology and biochemistry of plant cell walls. In Topics in Plant Physiology, M. Black and J. Chapman, eds (London: Unwin Hyman), pp. 6–57.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

