The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue
- PMID: 10810148
- PMCID: PMC139925
- DOI: 10.1105/tpc.12.5.757
The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue
Abstract
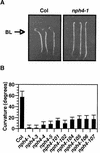
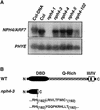
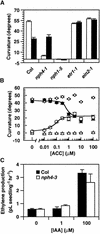
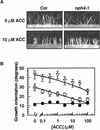
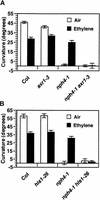
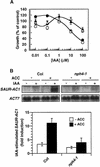
Organ bending through differential growth represents a major mechanism by which plants are able to adaptively alter their morphology in response to local changes in the environment. Two plant hormones, auxin and ethylene, have been implicated as regulators of differential growth responses; however, the mechanisms by which they elicit their effects remain largely unknown. Here, we describe isolation of the NPH4 gene of Arabidopsis, which is conditionally required for differential growth responses of aerial tissues, and we report that NPH4 encodes the auxin-regulated transcriptional activator ARF7. The phenotypes of nph4 mutants, which include multiple differential growth defects associated with reduced auxin responsiveness, including impaired auxin-induced gene expression, are consistent with the predicted loss of function of a transcriptional activator, and these phenotypes indicate that auxin-dependent changes in gene transcription are prerequisite for proper organ bending responses. Although NPH4/ARF7 appears to be a major regulator of differential growth, it is not the sole regulator because phenotypes of nph4 null mutants were suppressed by application of ethylene. This latter finding illustrates the intimate connection between auxin and ethylene in the control of growth in higher plants.
Figures


References
-
- Alonso, J.M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J.R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152. - PubMed
-
- Ausubel, F., Brent, R., Kingston, R.E., Moore, D.D, Seidman, J.G., Smith, J.A, and Struhl, K., eds (1995). Short Protocols in Molecular Biology. (New York: John Wiley).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

