Protein phosphatase 1alpha is a Ras-activated Bad phosphatase that regulates interleukin-2 deprivation-induced apoptosis
- PMID: 10811615
- PMCID: PMC384373
- DOI: 10.1093/emboj/19.10.2237
Protein phosphatase 1alpha is a Ras-activated Bad phosphatase that regulates interleukin-2 deprivation-induced apoptosis
Abstract
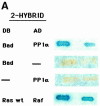
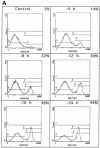
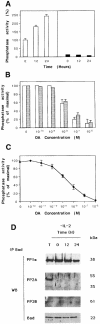
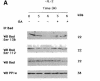
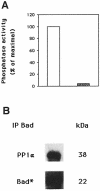
Growth factor deprivation is a physiological mechanism to regulate cell death. We utilize an interleukin-2 (IL-2)-dependent murine T-cell line to identify proteins that interact with Bad upon IL-2 stimulation or deprivation. Using the yeast two-hybrid system, glutathione S-transferase (GST) fusion proteins and co-immunoprecipitation techniques, we found that Bad interacts with protein phosphatase 1alpha (PP1alpha). Serine phosphorylation of Bad is induced by IL-2 and its dephosphorylation correlates with appearance of apoptosis. IL-2 deprivation induces Bad dephosphorylation, suggesting the involvement of a serine phosphatase. A serine/threonine phosphatase activity, sensitive to the phosphatase inhibitor okadaic acid, was detected in Bad immunoprecipitates from IL-2-stimulated cells, increasing after IL-2 deprivation. This enzymatic activity also dephosphorylates in vivo (32)P-labeled Bad. Treatment of cells with okadaic acid blocks Bad dephosphorylation and prevents cell death. Finally, Ras activation controls the catalytic activity of PP1alpha. These results strongly suggest that Bad is an in vitro and in vivo substrate for PP1alpha phosphatase and that IL-2 deprivation-induced apoptosis may operate by regulating Bad phosphorylation through PP1alpha phosphatase, whose enzymatic activity is regulated by Ras.
Figures









References
-
- Adams J.M. and Cory,S. (1998) The Bcl-2 protein family: arbiters of cell survival. Science, 281, 1322–1326. - PubMed
-
- Axton J.M., Dombradi,V., Cohen,P.T. and Glover,D.M. (1990) One of the protein phosphatase 1 isoenzymes in Drosophila is essential for mitosis. Cell, 63, 33–46. - PubMed
-
- Baharians Z. and Schönthal,A.H. (1999) Reduction of H-Ras induced cellular transformation by elevated expression of protein phosphatase type 2A. Mol. Carcinog., 24, 246–254. - PubMed
-
- Bartel P.L. and Fields,S. (1997) The Yeast Two-Hybrid System. Oxford University Press, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

