High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry
- PMID: 10811876
- PMCID: PMC18514
- DOI: 10.1073/pnas.090099097
High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry
Abstract
We have used intramolecular cross-linking, MS, and sequence threading to rapidly identify the fold of a model protein, bovine basic fibroblast growth factor (FGF)-2. Its tertiary structure was probed with a lysine-specific cross-linking agent, bis(sulfosuccinimidyl) suberate (BS(3)). Sites of cross-linking were determined by tryptic peptide mapping by using time-of-flight MS. Eighteen unique intramolecular lysine (Lys-Lys) cross-links were identified. The assignments for eight cross-linked peptides were confirmed by using post source decay MS. The interatomic distance constraints were all consistent with the tertiary structure of FGF-2. These relatively few constraints, in conjunction with threading, correctly identified FGF-2 as a member of the beta-trefoil fold family. To further demonstrate utility, we used the top-scoring homolog, IL-1beta, to build an FGF-2 homology model with a backbone error of 4.8 A (rms deviation). This method is fast, is general, uses small amounts of material, and is amenable to automation.
Figures
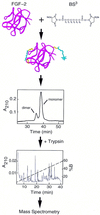

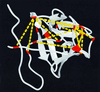

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

