Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli
- PMID: 10811894
- PMCID: PMC18519
- DOI: 10.1073/pnas.100118697
Domain organization and flavin adenine dinucleotide-binding determinants in the aerotaxis signal transducer Aer of Escherichia coli
Abstract
Aerotactic responses in Escherichia coli are mediated by the membrane transducer Aer, a recently identified member of the superfamily of PAS domain proteins, which includes sensors of light, oxygen, and redox state. Initial studies of Aer suggested that it might use a flavin adenine dinucleotide (FAD) prosthetic group to monitor cellular redox changes. To test this idea, we purified lauryl maltoside-solubilized Aer protein by His-tag affinity chromatography and showed by high performance liquid chromatography, mass spectrometry, and absorbance spectroscopy that it bound FAD noncovalently. Polypeptide fragments spanning the N-terminal 290 residues of Aer, which contains the PAS motif, were able to bind FAD. Fusion of this portion of Aer to the flagellar signaling domain of Tsr, the serine chemoreceptor, yielded a functional aerotaxis transducer, demonstrating that the FAD-binding portion of Aer is sufficient for aerosensing. Aerotaxis-defective missense mutants identified two regions, in addition to the PAS domain, that play roles in FAD binding. Those regions flank a central hydrophobic segment needed to anchor Aer to the cytoplasmic membrane. They might contact the FAD ligand directly or stabilize the FAD-binding pocket. However, their lack of sequence conservation in Aer homologs of other bacteria suggests that they play less direct roles in FAD binding. One or both regions probably also play important roles in transmitting stimulus-induced conformational changes to the C-terminal flagellar signaling domain to trigger aerotactic behavioral responses.
Figures





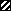 , NifL/PAS
similarity segment; IIII, F1 region; ▨, hydrophobic
segment; IIII, F2 region; ▨, MCP similarity
segment. (A) F1 and F2 directly comprise the FAD-binding
pocket. (B) F1 and F2 stabilize the FAD-binding pocket
through interactions with the NifL/PAS domain. (C) F1 and
F2 indirectly stabilize the FAD-binding pocket. In all three models,
input stimuli sensed by the FAD ligand, ostensibly via redox changes,
are transmitted to the output signaling domain through the F2 region.
The hydrophobic segment anchors Aer to the inner membrane, but
otherwise may play no role in input-output communication.
, NifL/PAS
similarity segment; IIII, F1 region; ▨, hydrophobic
segment; IIII, F2 region; ▨, MCP similarity
segment. (A) F1 and F2 directly comprise the FAD-binding
pocket. (B) F1 and F2 stabilize the FAD-binding pocket
through interactions with the NifL/PAS domain. (C) F1 and
F2 indirectly stabilize the FAD-binding pocket. In all three models,
input stimuli sensed by the FAD ligand, ostensibly via redox changes,
are transmitted to the output signaling domain through the F2 region.
The hydrophobic segment anchors Aer to the inner membrane, but
otherwise may play no role in input-output communication.References
-
- Armitage J P. Adv Microbiol Physiol. 1999;41:229–289. - PubMed
-
- Stock J B, Surette M G. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. I. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1103–1129.
-
- Blair D F. Annu Rev Microbiol. 1995;49:489–522. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

