Expression profiling of pancreatic beta cells: glucose regulation of secretory and metabolic pathway genes
- PMID: 10811900
- PMCID: PMC18509
- DOI: 10.1073/pnas.100126597
Expression profiling of pancreatic beta cells: glucose regulation of secretory and metabolic pathway genes
Abstract
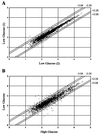
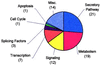
Pancreatic beta cells respond to changes in blood glucose by secreting insulin and increasing insulin synthesis. To identify genes used in these responses, we have carried out expression profiling of beta cells exposed to high (25 mM) or low (5.5 mM) glucose by using oligonucleotide microarrays. Functional clustering of genes that averaged a 2.2-fold or greater change revealed large groups of secretory pathway components, enzymes of intermediary metabolism, cell-signaling components, and transcription factors. Many secretory pathway genes were up-regulated in high glucose, including seven members of the endoplasmic reticulum (ER) translocon. In agreement with array analysis, protein levels of translocon components were increased by high glucose. Most dramatically, the alpha subunit of the signal recognition particle receptor was increased over 20-fold. These data indicate that the translocon and ribosome docking are major regulatory targets of glucose in the beta cell. Analysis of genes encoding enzymes of intermediary metabolism indicated that low glucose brought about greater utilization of amino acids as an energy source. This conclusion was supported by observations of increased urea production under low-glucose conditions. The above results demonstrate genome-wide integration of beta-cell functions at the level of transcript abundance and validate the efficacy of expression profiling in identifying genes involved in the beta-cell glucose response.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

