Secreted and membrane attractin result from alternative splicing of the human ATRN gene
- PMID: 10811918
- PMCID: PMC18552
- DOI: 10.1073/pnas.110139897
Secreted and membrane attractin result from alternative splicing of the human ATRN gene
Abstract
Attractin, initially identified as a soluble human plasma protein with dipeptidyl peptidase IV activity that is expressed and released by activated T lymphocytes, also has been identified as the product of the murine mahogany gene with connections to control of pigmentation and energy metabolism. The mahogany product, however, is a transmembrane protein, raising the possibility of a human membrane attractin in addition to the secreted form. The genomic structure of human attractin reveals that soluble attractin arises from transcription of 25 sequential exons on human chromosome 20p13, where the 3' terminal exon contains sequence from a long interspersed nuclear element-1 (LINE-1) retrotransposon element that includes a stop codon and a polyadenylation signal. The mRNA isoform for membrane attractin splices over the LINE-1 exon and includes five exons encoding transmembrane and cytoplasmic domains with organization and coding potential almost identical to that of the mouse gene. The relative abundance of soluble and transmembrane isoforms measured by reverse transcription-PCR is differentially regulated in lymphoid tissues. Because activation of peripheral blood leukocytes with phytohemagglutinin induces strong expression of cell surface attractin followed by release of soluble attractin, these results suggest that a genomic event unique to mammals, LINE-1 insertion, has provided an evolutionary mechanism for regulating cell interactions during an inflammatory reaction.
Figures

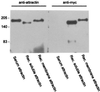


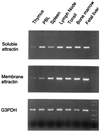
References
-
- Lider O, Hershkoviz R, Kachalsky S G. Crit Rev Immunol. 1995;15:271–283. - PubMed
-
- Duke-Cohan J S, Morimoto C, Rocker J A, Schlossman S F. J Immunol. 1996;156:1714–1721. - PubMed
-
- DeMeester I, Korom S, Van Damme J, Scharpe S. Immunol Today. 1999;20:367–375. - PubMed
-
- Gunn T M, Miller K A, He L, Hyman R W, Azarani A, Schlossman S F, Duke-Cohan J S, Barsh G S. Nature (London) 1999;398:152–156. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

