Interactions between heterologous forms of prion protein: binding, inhibition of conversion, and species barriers
- PMID: 10811921
- PMCID: PMC18520
- DOI: 10.1073/pnas.110523897
Interactions between heterologous forms of prion protein: binding, inhibition of conversion, and species barriers
Abstract
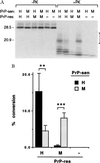
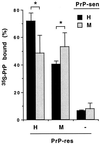
The self-induced formation of the disease-associated, protease-resistant prion protein (PrP-res) from the normal protease-sensitive isoform (PrP-sen) appears to be a key event in the pathogenesis of transmissible spongiform encephalopathies. The amino acid sequence specificity of PrP-res formation correlates with, and may account for, the species specificity in transmission of transmissible spongiform encephalopathy agents in vivo. To analyze the mechanism controlling the sequence specificity of PrP-res formation, we compared the binding of PrP-sen to PrP-res with its subsequent acquisition of protease resistance by using cell-free systems consisting of heterologous versus homologous mouse and hamster PrP isoforms. Our studies showed that heterologous PrP-sen can bind to PrP-res with little conversion to the protease-resistant state and, in doing so, can interfere with the conversion of homologous PrP-sen. The interference occurred with molar ratios of homologous to heterologous PrP-sen molecules as low as 1:1. The interference was due primarily to the inhibition of conversion, but not the binding, of the homologous PrP-sen to PrP-res. The results provide evidence that the sequence specificity of PrP-res formation in this model is determined more by the conversion to protease resistance than by the initial binding step. These findings also imply that after the initial binding, further intermolecular interactions between PrP-sen and PrP-res are required to complete the process of conversion to the protease-resistant state.
Figures




Similar articles
-
Specific binding of normal prion protein to the scrapie form via a localized domain initiates its conversion to the protease-resistant state.EMBO J. 1999 Jun 15;18(12):3193-203. doi: 10.1093/emboj/18.12.3193. EMBO J. 1999. PMID: 10369660 Free PMC article.
-
Glycosylation influences cross-species formation of protease-resistant prion protein.EMBO J. 2001 Dec 3;20(23):6692-9. doi: 10.1093/emboj/20.23.6692. EMBO J. 2001. PMID: 11726505 Free PMC article.
-
Sulfated glycans and elevated temperature stimulate PrP(Sc)-dependent cell-free formation of protease-resistant prion protein.EMBO J. 2001 Feb 1;20(3):377-86. doi: 10.1093/emboj/20.3.377. EMBO J. 2001. PMID: 11157745 Free PMC article.
-
Prion protein and species barriers in the transmissible spongiform encephalopathies.Biomed Pharmacother. 1999;53(1):27-33. doi: 10.1016/s0753-3322(99)80057-2. Biomed Pharmacother. 1999. PMID: 10221165 Review.
-
Formation of protease-resistant prion protein in cell-free systems.Curr Issues Mol Biol. 2000 Jul;2(3):95-101. Curr Issues Mol Biol. 2000. PMID: 11471561 Review.
Cited by
-
Efficient conversion of normal prion protein (PrP) by abnormal hamster PrP is determined by homology at amino acid residue 155.J Virol. 2001 May;75(10):4673-80. doi: 10.1128/JVI.75.10.4673-4680.2001. J Virol. 2001. PMID: 11312338 Free PMC article.
-
Identifying key components of the PrPC-PrPSc replicative interface.J Biol Chem. 2008 Dec 5;283(49):34021-8. doi: 10.1074/jbc.M804475200. Epub 2008 Sep 30. J Biol Chem. 2008. PMID: 18826953 Free PMC article.
-
Bank vole prion protein extends the use of RT-QuIC assays to detect prions in a range of inherited prion diseases.Sci Rep. 2021 Mar 4;11(1):5231. doi: 10.1038/s41598-021-84527-9. Sci Rep. 2021. PMID: 33664355 Free PMC article.
-
Dynamic interactions of Sup35p and PrP prion protein domains modulate aggregate nucleation and seeding.Prion. 2008 Jul-Sep;2(3):99-106. doi: 10.4161/pri.2.3.7147. Prion. 2008. PMID: 19195120 Free PMC article.
-
Heterozygous inhibition in prion infection: the stone fence model.Prion. 2009 Jan-Mar;3(1):27-30. doi: 10.4161/pri.3.1.8514. Epub 2009 Jan 23. Prion. 2009. PMID: 19372732 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

