CpG methylation as a mechanism for the regulation of E2F activity
- PMID: 10823896
- PMCID: PMC18629
- DOI: 10.1073/pnas.100340697
CpG methylation as a mechanism for the regulation of E2F activity
Abstract
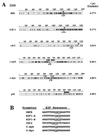
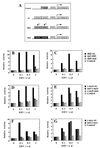
Regulation of gene expression in mammals through methylation of cytosine residues at CpG dinucleotides is involved in the development and progression of tumors. Because many genes that are involved in the control of cell proliferation are regulated by members of the E2F family of transcription factors and because some E2F DNA-binding sites are methylated in vivo, we have investigated whether CpG methylation can regulate E2F functions. We show here that methylation of E2F elements derived from the dihydrofolate reductase, E2F1, and cdc2 promoters prevents the binding of all E2F family members tested (E2F1 through E2F5). In contrast, methylation of the E2F elements derived from the c-myc and c-myb promoters minimally affects the binding of E2F2, E2F3, E2F4, and E2F5 but significantly inhibits the binding of E2F1. Consistent with these studies, E2F3, but not E2F1, activates transcription through methylated E2F sites derived from the c-myb and c-myc genes whereas both E2F1 and E2F3 fail to transactivate a reporter gene that is under the control of a methylated dihydrofolate reductase E2F site. Together, these data illustrate a means through which E2F activity can be controlled.
Figures



References
-
- Bird A P. Nature (London) 1986;321:209–213. - PubMed
-
- Cross S H, Bird A P. Curr Opin Genet Dev. 1995;5:309–314. - PubMed
-
- Cho K R, Hedrick L. Curr Top Microbiol Immunol. 1997;221:149–176. - PubMed
-
- Jones P A. Cancer Res. 1996;56:2463–2467. - PubMed
-
- Laird P W, Jaenisch R. Hum Mol Genet. 1994;3:1487–1495. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

