Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways
- PMID: 10823906
- PMCID: PMC18635
- DOI: 10.1073/pnas.110149597
Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways
Abstract
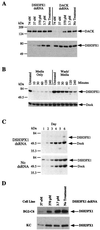
We demonstrate the efficacy of double-stranded RNA-mediated interference (RNAi) of gene expression in generating "knock-out" phenotypes for specific proteins in several Drosophila cell lines. We prove the applicability of this technique for studying signaling cascades by dissecting the well-characterized insulin signal transduction pathway. Specifically, we demonstrate that inhibiting the expression of the DSOR1 (mitogen-activated protein kinase kinase, MAPKK) prevents the activation of the downstream ERK-A (MAPK). In contrast, blocking ERK-A expression results in increased activation of DSOR1. We also show that Drosophila AKT (DAKT) activation depends on the insulin receptor substrate, CHICO (IRS1-4). Finally, we demonstrate that blocking the expression of Drosophila PTEN results in the activation of DAKT. In all cases, the interference of the biochemical cascade by RNAi is consistent with the known steps in the pathway. We extend this powerful technique to study two proteins, DSH3PX1 and Drosophila ACK (DACK). DSH3PX1 is an SH3, phox homology domain-containing protein, and DACK is homologous to the mammalian activated Cdc42 tyrosine kinase, ACK. Using RNAi, we demonstrate that DACK is upstream of DSH3PX1 phosphorylation, making DSH3PX1 an identified downstream target/substrate of ACK-like tyrosine kinases. These experiments highlight the usefulness of RNAi in dissecting complex biochemical signaling cascades and provide a highly effective method for determining the function of the identified genes arising from the Drosophila genome sequencing project.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

