Atomic force microscopy reveals the mechanical design of a modular protein
- PMID: 10823913
- PMCID: PMC18646
- DOI: 10.1073/pnas.120048697
Atomic force microscopy reveals the mechanical design of a modular protein
Abstract
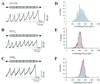
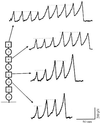
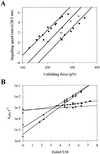
Tandem modular proteins underlie the elasticity of natural adhesives, cell adhesion proteins, and muscle proteins. The fundamental unit of elastic proteins is their individually folded modules. Here, we use protein engineering to construct multimodular proteins composed of Ig modules of different mechanical strength. We examine the mechanical properties of the resulting tandem modular proteins by using single protein atomic force microscopy. We show that by combining modules of known mechanical strength, we can generate proteins with novel elastic properties. Our experiments reveal the simple mechanical design of modular proteins and open the way for the engineering of elastic proteins with defined mechanical properties, which can be used in tissue and fiber engineering.
Figures

References
-
- Smith L B, Schaffer T E, Viani M, Thompson J B, Frederick N A, Kindt J, Belcher A, Studky G D, Morse D E, Hansma P K. Nature (London) 1999;399:761–763.
-
- Oberhauser A F, Marszalek P E, Erickson H P, Fernandez J M. Nature (London) 1998;393:181–185. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

