Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA
- PMID: 10823915
- PMCID: PMC18605
- DOI: 10.1073/pnas.120120397
Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA
Abstract
The extrinsic fluorescence dye 8-anilino-1-naphthalene sulfonate (ANS) is widely used for probing conformational changes in proteins, yet no detailed structure of ANS bound to any protein has been reported so far. ANS has been successfully used to monitor the induced-fit mechanism of MurA [UDPGlcNAc enolpyruvyltransferase (EC )], an essential enzyme for bacterial cell wall biosynthesis. We have solved the crystal structure of the ANS small middle dotMurA complex at 1.7-A resolution. ANS binds at an originally solvent-exposed region near Pro-112 and induces a major restructuring of the loop Pro-112-Pro-121, such that a specific binding site emerges. The fluorescence probe is sandwiched between the strictly conserved residues Arg-91, Pro-112, and Gly-113. Substrate binding to MurA is accompanied by large movements especially of the loop and Arg-91, which explains why ANS is an excellent sensor of conformational changes during catalysis of this pharmaceutically important enzyme.
Figures
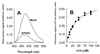
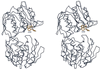
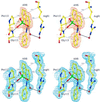
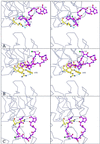
Comment in
-
Closing down on glyphosate inhibition--with a new structure for drug discovery.Proc Natl Acad Sci U S A. 2001 Mar 13;98(6):2944-6. doi: 10.1073/pnas.061025898. Proc Natl Acad Sci U S A. 2001. PMID: 11248008 Free PMC article. Review. No abstract available.
References
-
- Slavik J. Biochim Biophys Acta. 1982;694:1–25. - PubMed
-
- Semisotnov G V, Rodionova N A, Razgulyaev O I, Uversky V N, Gripas A F, Gilmanshin R I. Biopolymers. 1991;31:119–128. - PubMed
-
- Weber L D, Tulinsky A, Johnson J D, El-Bayoumi M A. Biochemistry. 1979;18:1297–1303. - PubMed
-
- Stryer L. J Mol Biol. 1965;13:482–495. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

