A short tandem repeat-based phylogeny for the human Y chromosome
- PMID: 10827105
- PMCID: PMC1287076
- DOI: 10.1086/302953
A short tandem repeat-based phylogeny for the human Y chromosome
Erratum in
- Am J Hum Genet 2000 Jul;67(1):270
Abstract
Human Y-chromosomal short tandem repeat (STR) data provide a potential model system for the understanding of autosomal STR mutations in humans and other species. Yet, the reconstruction of STR evolution is rarely attempted, because of the absence of an appropriate methodology. We here develop and validate a phylogenetic-network approach. We have typed 256 Y chromosomes of indigenous descent from Africa, Asia, Europe, Australia, and highland Papua New Guinea, for the STR loci DYS19, DXYS156Y, DYS389, DYS390, DYS392, and DYS393, as well as for five ancient biallelic mutation events: two poly (A) length variants associated with the YAP insertion, two independent SRY-1532 mutations, and the 92R7 mutation. We have used our previously published pedigree data from 11,000 paternity-tested autosomal STR-allele transfers to produce a two-class weighting system for the Y-STR loci that is based on locus lengths and motif lengths. Reduced-median-network analysis yields a phylogeny that is independently supported by the five biallelic mutations, with an error of 6%. We find the earliest branch in our African San (Bushmen) sample. Assuming an age of 20,000 years for the Native American DYS199 T mutation, we estimate a mutation rate of 2.6x10-4 mutations/20 years for slowly mutating Y STRs, approximately 10-fold slower than the published average pedigree rate.
Figures


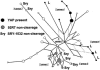

References
Electronic-Database Information
-
- Life Sciences and Engineering Technology Solutions, http://www.fluxus-engineering.com (for network methods)
References
-
- Bandelt H-J, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

