Association mapping in structured populations
- PMID: 10827107
- PMCID: PMC1287075
- DOI: 10.1086/302959
Association mapping in structured populations
Abstract
The use, in association studies, of the forthcoming dense genomewide collection of single-nucleotide polymorphisms (SNPs) has been heralded as a potential breakthrough in the study of the genetic basis of common complex disorders. A serious problem with association mapping is that population structure can lead to spurious associations between a candidate marker and a phenotype. One common solution has been to abandon case-control studies in favor of family-based tests of association, such as the transmission/disequilibrium test (TDT), but this comes at a considerable cost in the need to collect DNA from close relatives of affected individuals. In this article we describe a novel, statistically valid, method for case-control association studies in structured populations. Our method uses a set of unlinked genetic markers to infer details of population structure, and to estimate the ancestry of sampled individuals, before using this information to test for associations within subpopulations. It provides power comparable with the TDT in many settings and may substantially outperform it if there are conflicting associations in different subpopulations.
Figures

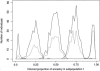
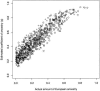
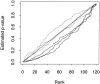
References
-
- Dempster AP, Laird NM, Rubin DB (1977) Maximum likelihood estimation from incomplete data via the EM algorithm (with discussion). J R Stat Soc B 39:1–38
-
- Devlin B, Roeder K (1999) Genomic control for association studies. Biometrics 55:997–1004 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

