Molecular and physiological responses of two classes of marine chromophytic phytoplankton (Diatoms and prymnesiophytes) during the development of nutrient-stimulated blooms
- PMID: 10831410
- PMCID: PMC110529
- DOI: 10.1128/AEM.66.6.2349-2357.2000
Molecular and physiological responses of two classes of marine chromophytic phytoplankton (Diatoms and prymnesiophytes) during the development of nutrient-stimulated blooms
Abstract
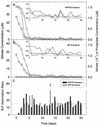
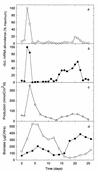
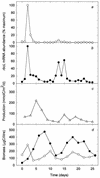
Generic taxon-specific DNA probes that target an internal region of the gene (rbcL) encoding the large subunit of ribulose-1, 5-bisphosphate carboxylase/oxygenase (RubisCO) were developed for two groups of marine phytoplankton (diatoms and prymnesiophytes). The specificity and utility of the probes were evaluated in the laboratory and also during a 1-month mesocosm experiment in which we investigated the temporal variability in RubisCO gene expression and primary production in response to inorganic nutrient enrichment. We found that the onset of successive bloom events dominated by each of the two classes of chromophyte algae was associated with marked taxon-specific increases in rbcL transcription rates. These observations suggest that measurements of RubisCO gene expression can provide an early indicator of the development of phytoplankton blooms and may also be useful in predicting which taxa are likely to dominate a bloom.
Figures


Similar articles
-
Phytoplankton-group specific quantitative polymerase chain reaction assays for RuBisCO mRNA transcripts in seawater.Mar Biotechnol (NY). 2007 Nov-Dec;9(6):747-59. doi: 10.1007/s10126-007-9027-z. Epub 2007 Aug 13. Mar Biotechnol (NY). 2007. PMID: 17694413
-
Diversity of the ribulose bisphosphate carboxylase/oxygenase form I gene (rbcL) in natural phytoplankton communities.Appl Environ Microbiol. 1997 Sep;63(9):3600-6. doi: 10.1128/aem.63.9.3600-3606.1997. Appl Environ Microbiol. 1997. PMID: 9293012 Free PMC article.
-
Deciphering the diversity and distribution of chromophytic phytoplankton in the Bohai Sea and the Yellow Sea via RuBisCO genes (rbcL).Mar Pollut Bull. 2022 Nov;184:114193. doi: 10.1016/j.marpolbul.2022.114193. Epub 2022 Oct 7. Mar Pollut Bull. 2022. PMID: 36209535
-
Carbon concentrating mechanisms in eukaryotic marine phytoplankton.Ann Rev Mar Sci. 2011;3:291-315. doi: 10.1146/annurev-marine-120709-142720. Ann Rev Mar Sci. 2011. PMID: 21329207 Review.
-
The potential for co-evolution of CO2-concentrating mechanisms and Rubisco in diatoms.J Exp Bot. 2017 Jun 1;68(14):3751-3762. doi: 10.1093/jxb/erx130. J Exp Bot. 2017. PMID: 28645158 Review.
Cited by
-
Nitrate/nitrite assimilation system of the marine picoplanktonic cyanobacterium Synechococcus sp. strain WH 8103: effect of nitrogen source and availability on gene expression.Appl Environ Microbiol. 2003 Dec;69(12):7009-18. doi: 10.1128/AEM.69.12.7009-7018.2003. Appl Environ Microbiol. 2003. PMID: 14660343 Free PMC article.
-
Sequencing effort dictates gene discovery in marine microbial metagenomes.Environ Microbiol. 2020 Nov;22(11):4589-4603. doi: 10.1111/1462-2920.15182. Epub 2020 Sep 29. Environ Microbiol. 2020. PMID: 32743860 Free PMC article.
-
Analysis of facultative lithotroph distribution and diversity on volcanic deposits by use of the large subunit of ribulose 1,5-bisphosphate carboxylase/oxygenase.Appl Environ Microbiol. 2004 Apr;70(4):2245-53. doi: 10.1128/AEM.70.4.2245-2253.2004. Appl Environ Microbiol. 2004. PMID: 15066819 Free PMC article.
-
The identification of three novel genes involved in the rapid-growth regulation in a marine diatom, Skeletonema costatum.Mar Biotechnol (NY). 2009 May-Jun;11(3):356-67. doi: 10.1007/s10126-008-9150-5. Epub 2008 Oct 8. Mar Biotechnol (NY). 2009. PMID: 18841415
-
Phytoplankton-group specific quantitative polymerase chain reaction assays for RuBisCO mRNA transcripts in seawater.Mar Biotechnol (NY). 2007 Nov-Dec;9(6):747-59. doi: 10.1007/s10126-007-9027-z. Epub 2007 Aug 13. Mar Biotechnol (NY). 2007. PMID: 17694413
References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1994.
-
- Bidle K D, Azam F. Accelerated dissolution of diatom silica by marine bacterial assemblages. Nature. 1999;397:508–512.
-
- Broecker W, Peng T H. Tracers in the sea. New York, N.Y: Lamont-Doherty Geological Observatory, Columbia University; 1982.
-
- Daugbjerg N, Andersen R A. Phylogenetic analyses of the rbcL sequences from haptophytes and heterokont algae suggest their chloroplasts are unrelated. Mol Biol Evol. 1997;14:1242–1251. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

